Genomic Islands in the Full-Genome Sequence of an NAD-Hemin-Independent Avibacterium paragallinarum Strain Isolated from Peru
Author details:
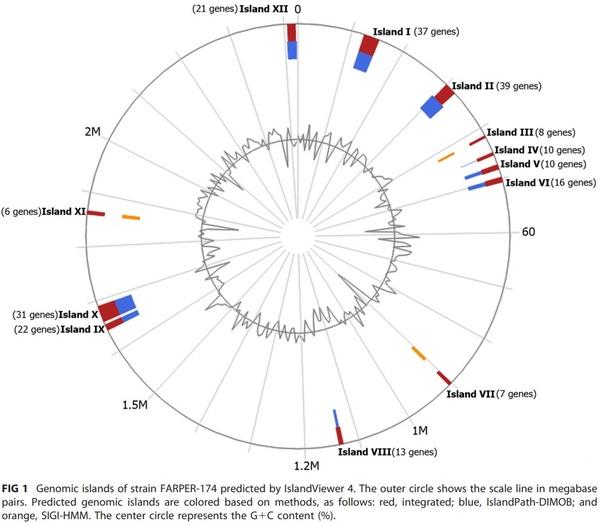
Here, we report the full-genome sequence of an NAD-hemin-independent Avibacterium paragallinarum serovar C-2 strain, FARPER-174, isolated from layer hens in Peru. This genome contained 12 potential genomic islands that include ribosomal protein-coding genes, a nadR gene, hemocin-coding genes, sequences of fagos, an rtx operon, and drug resistance genes.
1. Blackall PJ, Eaves LE, Rogers DG. 1990. Proposal of a new serovar and altered nomenclature for Haemophilus paragallinarum in the Kume hemagglutinin scheme. J Clin Microbiol 28:1185–1187.
2. Blackall PJ, Soriano-Vargas E. 2017. Infectious coryza and related bacterial infections, p. 859 – 873. In Swayne D (ed), Diseases of poultry. John
Wiley & Sons, Ltd, Chichester, United Kingdom.
3. Requena D, Chumbe A, Torres M, Alzamora O, Ramirez M, ValdiviaOlarte H, Gutierrez AH, Izquierdo-Lara R, Tataje-Lavanda L, Zavaleta
M, Tataje-Lavanda L, Best I, Fernández-Sánchez M, Icochea E, Zimic M,
Fernández-Díaz M, FARVET Research Group. 2013. Genome sequence and comparative analysis of Avibacterium paragallinarum. Bioinformation 9:528 –536. https://doi.org/10.6026/97320630009528.
4. Horta-Valerdi G, Sanchez-Alonso MP, Perez-Marquez VM, NegreteAbascal E, Vaca-Pacheco S, Hernandez-Gonzalez I, Gomez-Lunar Z,
Olmedo-Álvarez G, Vázquez-Cruz C. 2017. The genome sequence of
Avibacterium paragallinarum Strain CL has a large repertoire of insertion sequence elements. Genome Announc 5:e00152-17. https://doi.org/10
.1128/genomeA.00152-17.
5. Aguilar-Bultet L, Calderon-Copete SP, Frey J, Falquet L. 2013. Draft genome sequence of the virulent Avibacterium paragallinarum serotype
A strain JF4211 and identification of two toxins. Genome Announc
1:e00592-13. https://doi.org/10.1128/genomeA.00592-13.
6. Chen Y-C, Tan D-H, Shien J-H, Hsieh M-K, Yen T-Y, Chang P-C. 2014.
Identification and functional analysis of the cytolethal distending toxin gene from Avibacterium paragallinarum. Avian Pathol 43:43–50. https:// doi.org/10.1080/03079457.2013.861895.
7. Ho Sui SJ, Fedynak A, Hsiao WWL, Langille MGI, Brinkman FSL. 2009. The association of virulence factors with genomic islands. PLoS One 4:e8094. https://doi.org/10.1371/journal.pone.0008094.
8. Bertelli C, Tilley KE, Brinkman FSL. 2018. Microbial genomic island discovery, visualization and analysis. Brief Bioinform. https://doi.org/10
.1093/bib/bby042.
9. Dongsheng C, Hasan MS, Chen B. 2014. Identifying pathogenicity islands in bacterial pathogenomics using computational approaches. Pathogens
3:36 –56. https://doi.org/10.3390/pathogens3010036.
10. Falconi-Agapito F, Saravia LE, Flores-Pérez A, Fernández-Díaz M. 2015.
Naturally occurring -nicotinamide adenine dinucleotide–independent
Avibacterium paragallinarum isolate in Peru. Avian Dis 59:341–343. https://doi.org/10.1637/10969-110314-CaseR.
11. Blackall PJ, Christensen H, Beckenham T, Blackall LL, Bisgaard M. 2005.
Reclassification of Pasteurella gallinarum, [Haemophilus] paragallinarum,
Pasteurella avium and Pasteurella volantium as Avibacterium gallinarum gen. nov., comb. nov., Avibacterium paragallinarum comb. nov., Avibacterium avium comb. nov. and Avibacterium. Int J Syst Evol Microbiol
55:353–362. https://doi.org/10.1099/ijs.0.63357-0.
12. Chen X, Miflin JK, Zhang P, Blackall PJ. 1996. Development and application of DNA probes and PCR tests for Haemophilus paragallinarum. Avian
Dis 40:398 – 407. https://doi.org/10.2307/1592238.
13. Soriano VE, Blackall PJ, Dabo SM, Téllez G, García-Delgado GA, Fernández
RP. 2001. Serotyping of Haemophilus paragallinarum isolates from Mexico by the Kume hemagglutinin scheme. Avian Dis 45:680 – 683. https:// doi.org/10.2307/1592911.
14. Luna-Galaz GA, Morales-Erasto V, Peñuelas-Rivas CG, Blackall PJ, SorianoVargas E. 2016. Antimicrobial sensitivity of Avibacterium paragallinarum isolates from four Latin American countries. Avian Dis 60:673– 676. https://doi.org/10.1637/11398-022616-ResNote.1.
15. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual.
Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
16. Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum
A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013.
Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. https://doi.org/10.1038/ nmeth.2474.
17. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky
L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic
Genome Annotation Pipeline. Nucleic Acids Res 44:6614 –6624. https://doi
.org/10.1093/nar/gkw569.
18. Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing Group, Lau BY, Hoad G, Winsor GL, Brinkman F. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic
Acids Res 45:W30 –W35. https://doi.org/10.1093/nar/gkx343.












.jpg&w=3840&q=75)