Pathogen genomics and phage-based solutions for accurately identifying and controlling Salmonella pathogens
Author details:
Salmonella is a food-borne pathogen often linked to poultry sources, causing gastrointestinal infections in humans, with the numbers of multidrug resistant (MDR) isolates increasing globally. To gain insight into the genomic diversity of common serovars and their potential contribution to disease, we characterized antimicrobial resistance genes, and virulence factors encoded in 88 UK and 55 Thai isolates from poultry; the presence of virulence genes was detected through an extensive virulence determinants database compiled in this study. Long-read sequencing of three MDR isolates, each from a different serovar, was used to explore the links between virulence and resistance. To augment current control methods, we determined the sensitivity of isolates to 22 previously characterized Salmonella bacteriophages. Of the 17 serovars included, Salmonella Typhimurium and its monophasic variants were the most common, followed by S. Enteritidis, S. Mbandaka, and S. Virchow. Phylogenetic analysis of Typhumurium and monophasic variants showed poultry isolates were generally distinct from pigs. Resistance to sulfamethoxazole and ciprofloxacin was highest in isolates from the UK and Thailand, respectively, with 14–15% of all isolates being MDR. We noted that >90% of MDR isolates were likely to carry virulence genes as diverse as the srjF, lpfD, fhuA, and stc operons. Long-read sequencing revealed the presence of global epidemic MDR clones in our dataset, indicating they are possibly widespread in poultry. The clones included MDR ST198 S. Kentucky, harboring a Salmonella Genomic Island-1 (SGI)-K, European ST34 S. 1,4,[5],12:i:-, harboring SGI-4 and mercuryresistance genes, and a S. 1,4,12:i:- isolate from the Spanish clone harboring an MDR-plasmid. Testing of all isolates against a panel of bacteriophages showed variable sensitivity to phages, with STW-77 found to be the most effective. STW77 lysed 37.76% of the isolates, including serovars important for human clinical infections: S. Enteritidis (80.95%), S. Typhimurium (66.67%), S. 1,4,[5],12:i:- (83.3%), and S. 1,4,12: i:- (71.43%). Therefore, our study revealed that combining genomics and phage sensitivity assays is promising for accurately identifying and providing biocontrols for Salmonella to prevent its dissemination in poultry flocks and through the food chain to cause infections in humans.
KEYWORDS antimicrobial resistance, Salmonella, virulence genes, genomics, bacteriophages, serovar
Introduction
Materials and methods
Bacterial isolates and bacteriophages
Minimum inhibitory concentration determination
DNA extraction and short-read sequencing
Long-read sequencing
Phylogenetic and SNP analysis
Phage host range lytic assays
Results
![FIGURE 1 Maximum likelihood tree of S. Typhimurium, S. Typhimurium variant Copenhagen, S. 1,4,[5],12:i:-, and S. 1,4,12:i:- isolates based on SNPs in the coregenome. The tree was constructed in RaxML-NG using 17,263 single nucleotide polymorphisms (SNP). The isolate names have been colored based on their country, source, and database of origin. Dark blue corresponds to sequences from the APHA isolate collection belonging to the UK poultry included in this study (n = 21); dark green represents archived APHA UK pig isolates (n = 14); dark orange represents isolates from the study collection belonging to Thai poultry (n = 13); light blue represents Enterobase isolates belonging to UK poultry (n = 64); light green represents Enterobase isolates belonging to UK pigs (n = 96); light orange represents Enterobase isolates belonging to poultry from Thailand (n = 2); and light pink represents Enterobase isolates from pigs in Thailand (n = 57). Branches are colored in black for bootstrap values of > 70 to provide confidence. The outer ring indicates the serovar of the isolates.](/_next/image/?url=https%3A%2F%2Fimages.engormix.com%2FE_articles%2F54038_494.gif&w=1200&q=75)
Antimicrobial resistance characterization
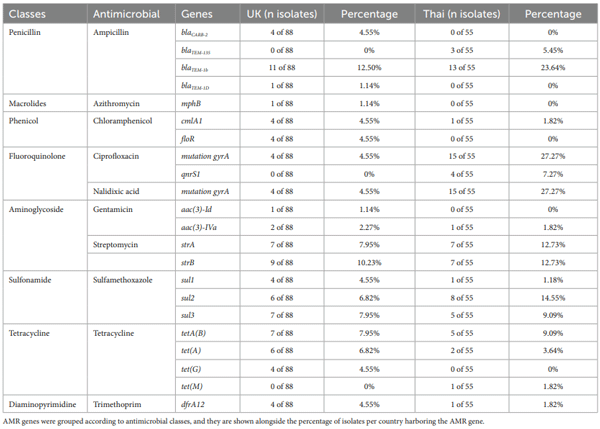
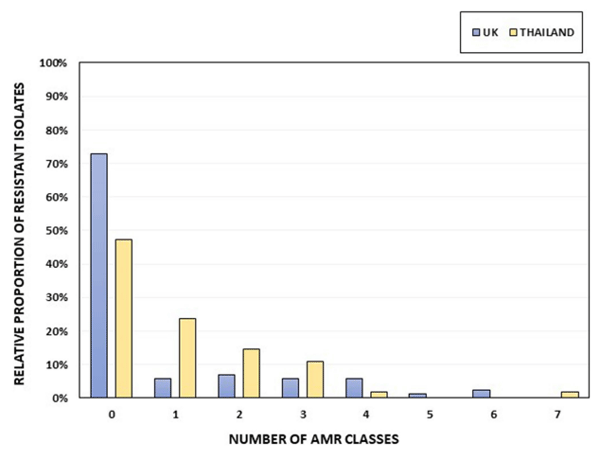
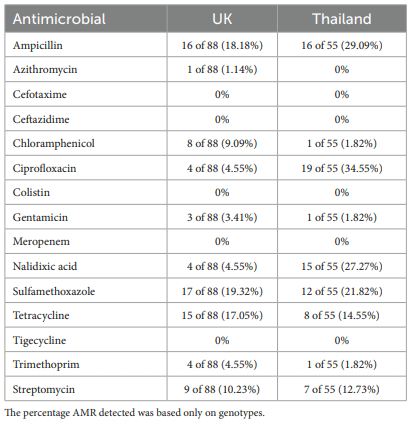
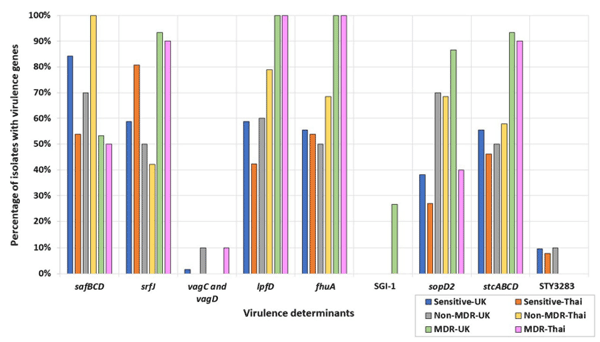
Virulence characterization
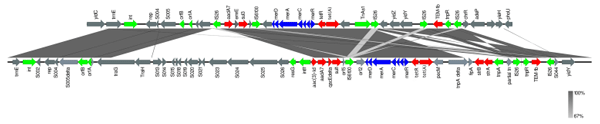
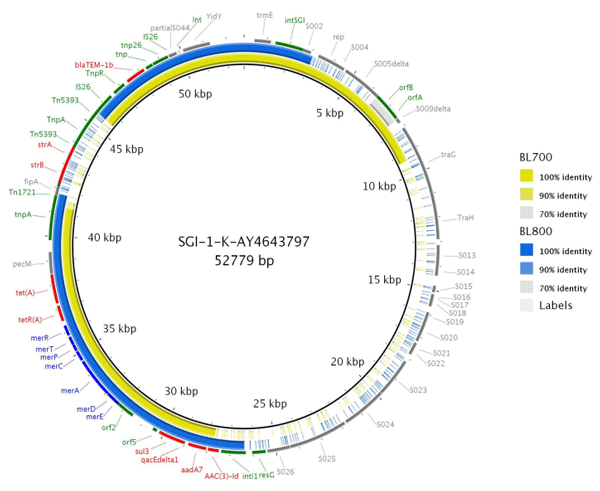
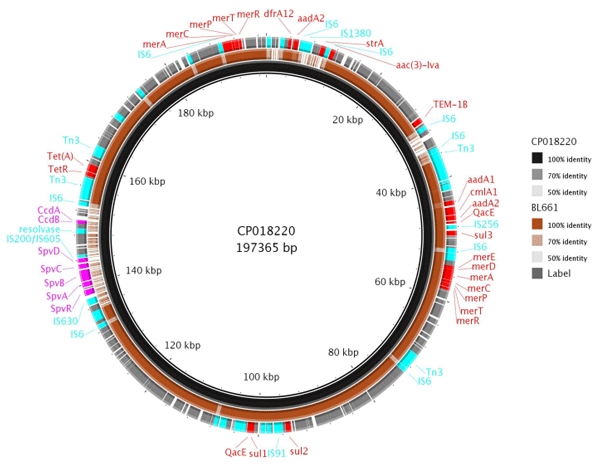
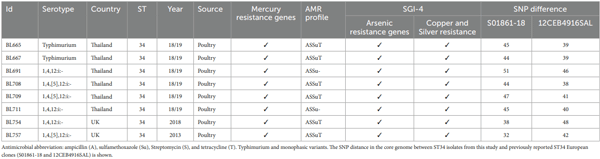
Phage lysis pattern for assessing host range
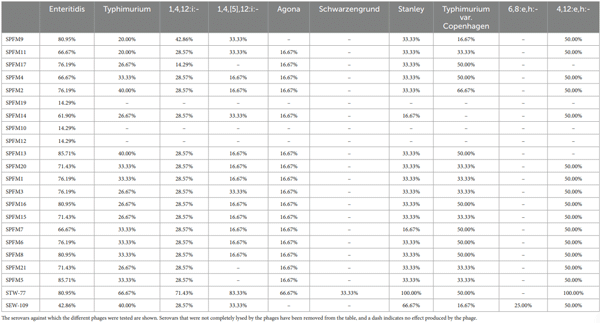
Discussion
Conclusion
Author’s summary
Data availability statement
Author contributions
Funding
Acknowledgments
Conflict of interest
Publisher’s note
Supplementary material
AbuOun, M., Jones, H., Stubberfield, E., Gilson, D., Shaw, L. P., Hubbard, A. T. M., et al. (2021). A genomic epidemiological study shows that prevalence of antimicrobial resistance in Enterobacterales is associated with the livestock host, as well as antimicrobial usage. Microb. Genom. 7. doi: 10.1099/mgen.0. 000630
AbuOun, M., O’Connor, H. M., Stubberfield, E. J., Nunez-Garcia, J., Sayers, E., Crook, D. W., et al. (2020). Characterizing antimicrobial resistant Escherichia coli and associated risk factors in a cross-sectional study of pig farms in Great Britain. Front. Microbiol. 11:861. doi: 10.3389/fmicb.2020.00861
Akil, L., and Ahmad, H. A. (2019). Quantitative risk assessment model of human salmonellosis resulting from consumption of broiler chicken. Diseases 7:19. doi: 10.3390/ diseases7010019
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
America IDSo (2011). Combating antimicrobial resistance: policy recommendations to save lives. Clin. Infect. Dis. 52, S397–S428. doi: 10.1093/cid/cir153
Antunes, P., Mourao, J., Pestana, N., and Peixe, L. (2011). Leakage of emerging clinically relevant multidrug-resistant Salmonella clones from pig farms. J. Antimicrob. Chemother. 66, 2028–2032. doi: 10.1093/jac/dkr228
Balasubramanian, R., Im, J., Lee, J.-S., Jeon, H. J., Mogeni, O. D., Kim, J. H., et al. (2019). The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccin. Immunother. 15, 1421–1426. doi: 10.1080/21645515. 2018.1504717
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant salmonella Typhimurium monophasic clone ST34. Microorganisms. 7:298. doi: 10.3390/microorganisms7090298
Cadel-Six, S., Cherchame, E., Douarre, P.-E., Tang, Y., Felten, A., Barbet, P., et al. (2021). The spatiotemporal dynamics and microevolution events that favored the success of the highly clonal multidrug-resistant monophasic Salmonella typhimurium circulating in Europe. Front. Microbiol. 12:651124. doi: 10.3389/ fmicb.2021.651124
Card, R., Vaughan, K., Bagnall, M., Spiropoulos, J., Cooley, W., Strickland, T., et al. (2016). Virulence characterisation of Salmonella enterica isolates of differing antimicrobial resistance Recovered from UK livestock and imported meat samples. Front. Microbiol. 7:640. doi: 10.3389/fmicb.2016.00640
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/ nar/gki008
Chuanchuen, R., Pathanasophon, P., Khemtong, S., Wannaprasat, W., and Padungtod, P. (2008). Susceptibilities to antimicrobials and disinfectants in Salmonella isolates obtained from poultry and swine in Thailand. J. Vet. Med. Sci. 70, 595–601. doi: 10.1292/jvms.70.595
Cohen, E., Azriel, S., Auster, O., Gal, A., Zitronblat, C., Mikhlin, S., et al. (2021). Pathoadaptation of the passerine-associated Salmonella enterica serovar Typhimurium lineage to the avian host. PLoS Pathog. 17:e1009451. doi: 10.1371/journal.ppat.1009451
Cordero-Alba, M., Bernal-Bayard, J., and Ramos-Morales, F. (2012). SrfJ, a salmonella type III secretion system effector regulated by PhoP, RcsB, and IolR. J. Bacteriol. 194, 4226–4236. doi: 10.1128/JB.00173-12
Czaplewski, L., Bax, R., Clokie, M., Dawson, M., Fairhead, H., Fischetti, V. A., et al. (2016). Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251. doi: 10.1016/S1473-3099(15)00466-1
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
Department for Environment Food and Rural Affairs (2022). Monthly statistics on the activity of UK hatcheries and UK poultry slaughterhouses (data for August 2022) 22 September 2022, Available at: https://www.gov.uk/government/statistics/poultry-andpoultry-meat-statistics/monthly-statistics-on-the-activity-of-uk-hatcheries-and-ukpoultry-slaughterhouses-data-for-august-2022. (Accessed October 2022).
Directorate VM (2014). UK veterinary antibiotic resistance and sales surveillance. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/ uploads/attachment_data/file/477788/Optimised_version_-_VARSS_Report_2014__ Sales___Resistance_.pdf.
Duggett, N., AbuOun, M., Randall, L., Horton, R., Lemma, F., Rogers, J., et al. (2020). The importance of using whole genome sequencing and extended spectrum betalactamase selective media when monitoring antimicrobial resistance. Sci. Rep. 10:19880. doi: 10.1038/s41598-020-76877-7
Duggett, N. A., Randall, L. P., Horton, R. A., Lemma, F., Kirchner, M., Nunez-Garcia, J., et al. (2018). Molecular epidemiology of isolates with multiple mcr plasmids from a pig farm in Great Britain: the effects of colistin withdrawal in the short and long term. J. Antimicrob. Chemother. 73, 3025–3033. doi: 10.1093/jac/dky292
Duggett, N. A., Sayers, E., AbuOun, M., Ellis, R. J., Nunez-Garcia, J., Randall, L., et al. (2016). Occurrence and characterization of mcr-1-harbouring Escherichia coli isolated from pigs in Great Britain from 2013 to 2015. J. Antimicrob. Chemother. 72, 691–695. doi: 10.1093/jac/dkw477
Duprilot, M., Decre, D., Genel, N., Drieux, L., Sougakoff, W., and Arlet, G. (2017). Diversity and functionality of plasmid-borne VagCD toxin–antitoxin systems of Klebsiella pneumoniae. J. Antimicrob. Chemother. 72, 1320–1326. doi: 10.1093/jac/ dkw569
Eguale, T., Gebreyes, W. A., Asrat, D., Alemayehu, H., Gunn, J. S., and Engidawork, E. (2015). Non-typhoidal Salmonella serotypes, antimicrobial resistance and co-infection with parasites among patients with diarrhea and other gastrointestinal complaints in Addis Ababa, Ethiopia. BMC Infect. Dis. 15, 497–499. doi: 10.1186/s12879-015-1235-y
Elnekave, E., Hong, S., Mather, A. E., Boxrud, D., Taylor, A. J., Lappi, V., et al. (2018). Salmonella enterica serotype 4,[5], 12: i:-in swine in the United States Midwest: an emerging multidrug-resistant clade. Clin. Infect. Dis. 66, 877–885. doi: 10.1093/cid/ cix909 EUCAST (2020). MIC EUCAST. Available at: https://mic.eucast.org/
Figueiredo, R., Card, R., Nunes, C., AbuOun, M., Bagnall, M. C., Nunez, J., et al. (2015). Virulence characterization of Salmonella enterica by a new microarray: detection and evaluation of the Cytolethal distending toxin gene activity in the unusual host S. typhimurium. PLoS One 10:e0135010. doi: 10.1371/journal.pone.0135010
Foley, S., Lynne, A., and Nayak, R. (2008). Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 86, E149–E162. doi: 10.2527/jas.2007-0464
García, P., Guerra, B., Bances, M., Mendoza, M. C., and Rodicio, M. R. (2011). IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging salmonella enterica serotype 4,[5], 12: i:−. J. Antimicrob. Chemother. 66, 543–549. doi: 10.1093/jac/dkq481
Grimont, PA, and Weill, F-X (2007). Antigenic formulae of the Salmonella serovars. WHO collaborating centre for reference and research on Salmonella. 9:1–166.
Han, J., Lynne, A. M., David, D. E., Nayak, R., and Foley, S. L. (2012). Sequencing of plasmids from a multi-antimicrobial resistant Salmonella enterica serovar Dublin strain. Food Res. Int. 45, 931–934. doi: 10.1016/j.foodres.2011.04.016
Hawkey, J., Le Hello, S., Doublet, B., Granier, S. A., Hendriksen, R. S., Fricke, W. F., et al. (2019). Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb. Genom. 5. doi: 10.1099/mgen.0.000269
Hengkrawit, K., and Tangjade, C. (2022). Prevalence and trends in antimicrobial susceptibility patterns of multi-drug-resistance non-typhoidal Salmonella in Central Thailand, 2012–2019. Infect. Drug Resist. 15, 1305–1315. doi: 10.2147/IDR.S355213
Holman, D. B., Bearson, S. M. D., Bearson, B. L., and Brunelle, B. W. (2018). Chlortetracycline and florfenicol induce expression of genes associated with pathogenicity in multidrug-resistant Salmonella enterica serovar Typhimurium. Gut pathog. 10:10. doi: 10.1186/s13099-018-0236-y
Ingle, D. J., Ambrose, R. L., Baines, S. L., Duchene, S., Gonçalves da Silva, A., Lee, D. Y., et al. (2021). Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,[5], 12: i:-in Australia. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-021-25073-w
Jain, P., Chowdhury, G., Samajpati, S., Basak, S., Ganai, A., Samanta, S., et al. (2020). Characterization of non-typhoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000-2016. Braz. J. Microbiol. 51, 613–627. doi: 10.1007/s42770-019-00213-z
Jiang, X., Rossanese, O. W., Brown, N. F., Kujat-Choy, S., Galán, J. E., Finlay, B. B., et al. (2004). The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol. Microbiol. 54, 1186–1198. doi: 10.1111/j.1365-2958.2004.04344.x
Jung, L.-s., Ding, T., and Ahn, J. (2017). Evaluation of lytic bacteriophages for control of multidrug-resistant salmonella Typhimurium. Ann. Clin. Microbiol. Antimicrob. 16, 1–9. doi: 10.1186/s12941-017-0237-6
Kogut, M. H., and Arsenault, R. J. (2017). Immunometabolic phenotype alterations associated with the induction of disease tolerance and persistent asymptomatic infection of Salmonella in the chicken intestine. Front. Immunol. 8:372. doi: 10.3389/ fimmu.2017.00372
Kombade, S., and Kaur, N. (2021). Pathogenicity Island in Salmonella. Salmonella spp-a global challenge IntechOpen. doi: 10.5772/intechopen.96443
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxMLNG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Le Hello, S., Hendriksen, R. S., Doublet, B., Fisher, I., Nielsen, E. M., Whichard, J. M., et al. (2011). International spread of an epidemic population of salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204, 675–684. doi: 10.1093/infdis/jir409
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, Z., Ma, W., Li, W., Ding, Y., Zhang, Y., Yang, Q., et al. (2020). A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 132:109011. doi: 10.1016/j.foodres.2020.109011
Liljebjelke, K. A., Hofacre, C. L., White, D. G., Ayers, S., Lee, M. D., and Maurer, J. J. (2017). Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial poultry farms. Front. Vet. Sci. 4:96. doi: 10.3389/fvets.2017.00096
Lynch, J. P.III, Clark, N. M., and Zhanel, G. G. (2013). Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum β-lactamases and carbapenemases). Expert. Opin. Pharmacother. 14, 199–210. doi: 10.1517/14656566.2013.763030
Machado Junior, P. C., Chung, C., and Hagerman, A. (2020). Modeling Salmonella spread in broiler production: identifying determinants and control strategies. Front. Vet. Sci. 7:564. doi: 10.3389/fvets.2020.00564
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O'Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Nale, D. J., Ahmed, B., Haigh, R., Shan, J., Phothaworn, P., Thiennimitr, P., et al. (2023). Activity of a bacteriophage cocktail to control Salmonella growth ex vivo in avian, porcine, and human epithelial cell cultures. Phage 4, 11–25. doi: 10.1089/ phage.2023.0001
Nale, J. Y., Vinner, G. K., Lopez, V. C., Thanki, A. M., Phothaworn, P., Thiennimitr, P., et al. (2021). An optimized bacteriophage cocktail can effectively control Salmonella in vitro and in Galleria mellonella. Front. Microbiol. 11:609955. doi: 10.3389/ fmicb.2020.609955
Nobrega, F. L., Costa, A. R., Kluskens, L. D., and Azeredo, J. (2015). Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23, 185–191. doi: 10.1016/j.tim.2015.01.006
Nunez-Garcia, J., AbuOun, M., Storey, N., Brouwer, M. S., Delgado-Blas, J. F., Mo, S. S., et al. (2022). Harmonisation of in-silico next-generation sequencing based methods for diagnostics and surveillance. Sci. Rep. 12:14372. doi: 10.1038/s41598-022-16760-9
O’Neil, J. (2014). Review on antibiotic resisitance. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations Heal Wealth Nations. 1–16.
Pan, Z., Carter, B., Núñez-García, J., AbuOun, M., Fookes, M., Ivens, A., et al. (2009). Identification of genetic and phenotypic differences associated with prevalent and non-prevalent Salmonella enteritidis phage types: analysis of variation in amino acid transport. Microbiology 155, 3200–3213. doi: 10.1099/mic.0. 029405-0
Patchanee, P., Tanamai, P., Tadee, P., Hitchings, M. D., Calland, J. K., Sheppard, S. K., et al. (2020). Whole-genome characterisation of multidrug resistant monophasic variants of Salmonella typhimurium from pig production in Thailand. PeerJ 8:e9700. doi: 10.7717/peerj.9700
Pawar, S., Ashraf, M., Mehata, K., and Lahiri, C. (2017). Computational identification of indispensable virulence proteins of Salmonella typhi CT18. Curr. Top. Salmonella Salmonellosis. doi: 10.5772/66489 Available at https://www.intechopen.com/ chapters/53363
Petrovska, L., Mather, A. E., AbuOun, M., Branchu, P., Harris, S. R., Connor, T., et al. (2016). Microevolution of monophasic Salmonella typhimurium during epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. 22, 617–624. doi: 10.3201/ eid2204.150531
Phothaworn, P., Supokaivanich, R., Lim, J., Klumpp, J., Imam, M., Kutter, E., et al. (2020). Development of a broad-spectrum SALMONELLA phage cocktail containing Viunalike and Jerseylike viruses isolated from Thailand. Food Microbiol. 92:103586. doi: 10.1016/j.fm.2020.103586
Pullinger, G. D., and Lax, A. J. (1992). A salmonella Dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 6, 1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x
Romero-Calle, D., Guimarães Benevides, R., Góes-Neto, A., and Billington, C. (2019). Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics 8:138. doi: 10.3390/antibiotics8030138
Salmond, G. P., and Fineran, P. C. (2015). A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786. doi: 10.1038/nrmicro3564
Schroeder, M., Brooks, B. D., and Brooks, A. E. (2017). The complex relationship between virulence and antibiotic resistance. Genes 8:39. doi: 10.3390/genes8010039
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T. (2015). Snippy: Fast bacterial variant calling from NGS reads. Service USDoAFA (2021). Poultry and products annual: United States Department of Agriculture. Foreign Agricultural Service. Available at: https://apps. fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=Pou ltry%20and%20Products%20Annual_Bangkok_Thailand_09-01-2021.pdf. (Accessed August 2022).
Stubberfield, E., AbuOun, M., Sayers, E., O'Connor, H. M., Card, R. M., and Anjum, M. F. (2019). Use of whole genome sequencing of commensal Escherichia coli in pigs for antimicrobial resistance surveillance, United Kingdom, 2018. Euro Surveill. 24. doi: 10.2807/1560-7917.ES.2019.24.50.1900136
Sukjoi, C., Buddhasiri, S., Tantibhadrasapa, A., Kaewsakhorn, T., Phothaworn, P., Nale, J. Y., et al. (2022). Therapeutic effects of oral administration of lytic salmonella phages in a mouse model of non-typhoidal salmonellosis. Front. Microbiol. 13:13. doi: 10.3389/fmicb.2022.955136
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Thanki, A. M., Brown, N., Millard, A. D., and Clokie, M. R. (2019). Genomic characterization of jumbo salmonella phages that effectively target United Kingdom pig-associated Salmonella serotypes. Front. Microbiol. 10:1491. doi: 10.3389/ fmicb.2019.01491
Thermo Fisher Scientific (n.d.). Antimicrobial susceptibility testing. Available at https://www.thermofisher.com/uk/en/home/clinical/clinical-microbiology/ antimicrobial-susceptibility-testing.html
Veterinary Medicines Directorate (2021). UK veterinary antibiotic resistance and sales surveillance report 2020 2021. Available at: https://assets.publishing.service. gov.uk/government/uploads/system/uploads/attachment_data/ file/1072796/03.05.22_VARSS_Main_Report__Final_Accessible_version__3_.pdf. (Accessed October 2022)
Vikesland, P., Garner, E., Gupta, S., Kang, S., Maile-Moskowitz, A., and Zhu, N. (2019). Differential drivers of antimicrobial resistance across the world. Acc. Chem. Res. 52, 916–924. doi: 10.1021/acs.accounts.8b00643
Wales, A. D., Allen, V. M., and Davies, R. H. (2010). Chemical treatment of animal feed and water for the control of salmonella. Foodborne Pathog. Dis. 7, 3–15. doi: 10.1089/fpd.2009.0373
Wang, Y., Chen, X., Hu, Y., Zhu, G., White, A. P., and Köster, W. (2018). Evolution and sequence diversity of FhuA in Salmonella and Escherichia. Infect. Immun. 86. doi: 10.1128/IAI.00573-18
Weinberger, M., and Keller, N. (2005). Recent trends in the epidemiology of nontyphoid Salmonella and antimicrobial resistance: the Israeli experience and worldwide review. Curr. Opin. Infect. Dis. 18, 513–521. doi: 10.1097/01. qco.0000186851.33844.b2
Wen, J., Gou, H., Zhan, Z., Gao, Y., Chen, Z., Bai, J., et al. (2020). A rapid novel visualized loop-mediated isothermal amplification method for salmonella detection targeting at fimW gene. Poult. Sci. 99, 3637–3642. doi: 10.1016/j. psj.2020.03.045
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wongsuvan, G., Wuthiekanun, V., Hinjoy, S., Day, N. P., and Limmathurotsakul, D. (2018). Antibiotic use in poultry: a survey of eight farms in Thailand. Bull. World Health Organ. 96, 94–100. doi: 10.2471/BLT.17.195834
Zeng, L., Zhang, L., Wang, P., and Meng, G. (2017). Structural basis of host recognition and biofilm formation by salmonella Saf pili. elife:6. doi: 10.7554/eLife.28619
Zhang, L., Fu, Y., Xiong, Z., Ma, Y., Wei, Y., Qu, X., et al. (2018). Highly prevalent multidrug-resistant salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 9:2104. doi: 10.3389/fmicb.2018.02104









.jpg&w=3840&q=75)