Viral Disease in Swine: An Immunologist’s Perspective with Focus on PRRS
![Ontogeny of lymphocytes development from progenitor cells to virus-specific B and T cells. Pathways include those genomes encoded (green brackets), events stimulated by encounter with receptors of the innate immune system (orange brackets) and events that are somatically generated and cannot be passed to the offspring (blue brackets). [Reference Table 3]](/_next/image/?url=https%3A%2F%2Fimages.engormix.com%2FE_articles%2F0_120.gif&w=1200&q=75)
Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D Chladek, D. Characterization of swine infertility and respiratory syndrome (SIRS)virus (isolate ATCC VR-2332). J Vet Diagn Invest, 4:127-33, 1992.
Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol, 3:169-176, 2003
Brockmeier SL, Loving CL, Nelson EA, Miller LC, Nicholson TL, Register KB, Grubman MJ, Brough DE, Kehrli ME Jr. The presence of alpha interferon at the time of infection alters the innate and adaptive immune responses to porcine reproductive and respiratory syndrome virus. Clin Vaccine Immunol, 9:508-14,2010.
Buddaert W, Van Reeth K, Pensaert M. In vivo and in vitro 2017 interferon (IFN) studies with the porcine reproductive and 2018 respiratory syndrome virus (PRRSV). Adv Exp Med Biol, 440:461-7. 1998.
Butler JE, Lager, KM, Golde, W, Faaberg, KS, Sinkora, M, Loving, C, Zhang, YI. Porcine reproductive and respiratory syndrome (PRRS): an immune dysregulatory pandemics. Immunol. Res, 59:81-108, 2014.
Butler JE, Lemke CD, Weber P, Sinkora M, Lager KM. Antibody repertoire development in fetal and neonatal piglets. XIX. Undiversified B cells with hydrophobic HCDR3s preferentially proliferate in PRRS. J Immunol, 178:6320-31,2007.
Butler JE, Weber P, Sinkora M, Baker D, Schoenherr A, Mayer B, Francis D. Antibody repertoire development in fetal and neonatal piglets. VIII. Colonization is required for newborn piglets to make serum antibodies to T-dependent and type 2 T-independent antigens. J. Immunol, 169:6822-30, 2002.
Butler JE, Weber P, Wertz N, Lager KM. Porcine reproductive and respiratory syndrome virus (PRRSV) subverts development of adaptive immunity by proliferation of germline-encoded B cells with hydrophobic HCDR3s. J Immunol, 180:2347-56,2008.
Butler JE, Wertz N, Sinkora M. Antibody repertoire development in swine. Annul. Rev. Animal Biosciences, 5:255-279, 2017.
Butler JE, Wertz N, Sun J, Sacco, RE. Comparison of the expressed porcine Vβ and Jβ repertoire of thymocytes and peripheral T cells. Immunology 114:184-193,2005.
Butler JE. Transmission of immunity from mother to young. pp. 92-98. T. Hasegawa,M. Hayasbi, F. J. G., Ebling, and I. W. Henderson [Eds.], Fertility and Sterility. Exerpta Medica, Amsterdam, pp 92-98,1971.
Butler JE, Francis D, Freeling J, Weber P, Sun J, Krieg AM. Antibody repertoire development in fetal and neonatal piglets. IX. Three PAMPs act synergistically to allow germfree piglets to respond to TI-2 and TD antigens. J. Immunol, 175:6772-6785, 2005.
Butler JE, Sinkora M Wertz N, Holtmeier W, Lemke KD. Development of the neonatal B- and T-cell repertoire in swine: Implications for comparative and veterinary immunology. In: Mucosal immunity in domestic animals (K. Haverson, M. Bailey, B. Charley, Guest Eds) Veterinary Research, 37:417-441, 2006.
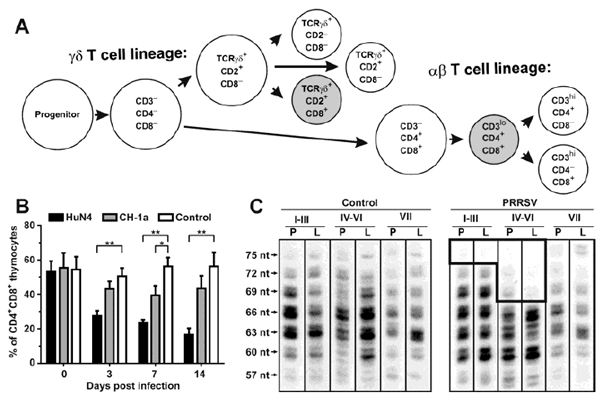
Butler JE, Sinkora M, Wang G, Stepanova K, Li Y, Cai. X Perturbation of thymocyte development underlies the PRRS pandemic: A testable hypothesis. Frontiers in Immunol, doi:10.389/fimmu 201.01077, 2019.
Calzada-Nova G, Schnitzlein WM, Husmann, RJ, Zuckermann, FA. North American porcine reproductive and respiratory syn2027 drome viruses inhibit type I interferon production by plasma2028 cytoid dendritic cells. J Virol, 2011:85:2703-13, 2006.
Cerottini J-C, McConahey PJ, Dixon FJ. Specificity of the immunosuppression caused by passive administration of antibody. J. Immunol, 103:268-275.
Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek Dan. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest, 4:117-26,1992.
Done SH, Paton, DJ. Porcine reproductive and respiratory syndrome: clinical disease, pathology and immunosuppression. Vet Rec, 136:32-35,1995.
Faaberg KS, Balasuriya UB, Brinton MA, Gorbalenya AE, Leung FC, Nauwynck H, Snijder EJ, Stadejek T, Yang H, Yoo D. Family Arteriviridae. In: King AMQ, editor. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier/Academic Press;. p.796-805, 2012.
Giradin SE, Hugot J-P, Samonetti, PJ. Lessons from NOD2 studies: toward a link between Crohn’s disease and bacterial sensing. Trends Immunol, 24:652-658, 2003
Heyman B. Feedback regulation of IgG antibodies. Immunology Letters, 88:157-161, 2003.
Hoffmann JA. Innate immunity, Curr Opin Immunol, 7:4-10, 1995.
Holtkamp DJ, Kliebenstein, JB Neuman, EJ Zimmerman, JJ Piolto, HT Yoda, TK Wang, C Yeske, PE Mower, CL, Haley, CA. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health and Production, 21:72-84, 2013.
Joklik WK, Willett HP. Zinsser Microbiology, 16th Ed. Appleton Century Croft, New York p 2-7, 1968
Klobasa F, Butler JE, Habe F. Maternal-neonatal immunoregulation: Suppression of de novo immunoglobulin synthesis of IgG and IgA, but not IgM, in neonatal piglets by bovine colostrum, is lost upon storage. Amer J Vet Res, 51:1407-1412,1990.
Klobasa F, Werhahn E, Butler, JE. Regulation of humoral immunity in the piglet by immunoglobulins of maternal origin. Res Vet Sci, 31:195-206,1981.
Lager KM, Buckley AC Porcine antiviral immunity: How important is it? Frontiers in Immunology doi: 10.3389/fimmu, 2019.02258, 2019.
Lemke CD, Joseph SH, Spaete R, Adolphson D, Vorwald A, Lager K, Butler JE. Lymphoid hyperplasia resulting in immune dysregulation is caused by porcine reproductive and respiratory syndrome virus infection in neonatal pigs. J Immunol, 172:1916-25, 2004.
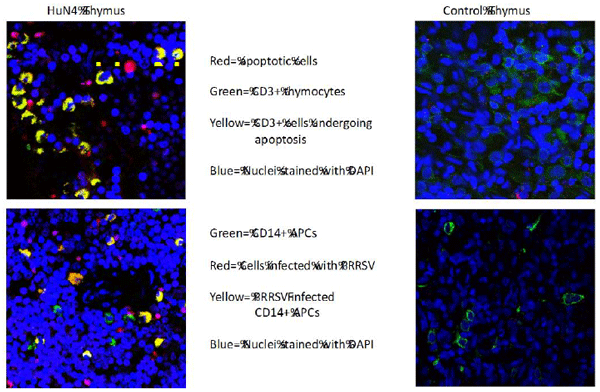
Li Y, Wang G, Liu Y, Tu Y, He Y, Wang Z , Han Z, Li L, Li A, Tao Y, Cai X. Identification of apoptotic cells in the thymus of piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Virus Research, 189:29-33,2014.
Lopez OJ, Oliveira MF, Alvarez-Garcia E, Kwon BJ, Doster A, Osoria FA. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clin Vac Immunol, 14:269-75, 2007.
Lu G, Pan J, Zhang, G. African swine fever virus in Asia: Its rapid spread and potential threat to unaffected countries. J Infection, doi: 10.1016/jinf 2019.11.011, 2019.
Mahapatra M, Parida S. Foot and mouth disease vaccine selection: current approaches and future perspectives. Expt. Rev. Vaccines, 17:577-591, 2018
Patel D, Nan Y, Shen M, Ritthipichai K, Zhu X, Zhang YJ. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol, 84:11045-55, 2010.
Renukaradhya GJ, Alekseev K, Jong K, Fang Y, Saif LJ. Porcine reproductive and respiratory syndrome virusinduced suppression exacerbates the inflammatory response to porcine respiratory coronavirus in pigs. Viral Immunol, 23:457-66, 2010.
Rowland RRR, Lawson S, Rossow K, Benfield, DA. Lymphotropism of porcine reproductive and respiratory syndrome virus replication during persistent infection of pigs originally exposed to virus in utero. Vet Microbiol, 96:219-235, 2003.
Sandez-Cordon PJ, Montoya M, Reis AL, Dixon LK. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet J, 233:41-48, 2018.
Sazzad HMS, Hossain MJ, Gurley ES, Ameen KMH, Parveen S, Islam MS, Faruque LI, Podder G, Banu SS, Lo MK, Rollin PE, Rota PA, Daszak P, Rahman M, Luby SP. Nipah virus infection outbreaks with nosocomial and corpse-to-human transmission, Bangladesh, Emerg Infect Dis, 19:210-217
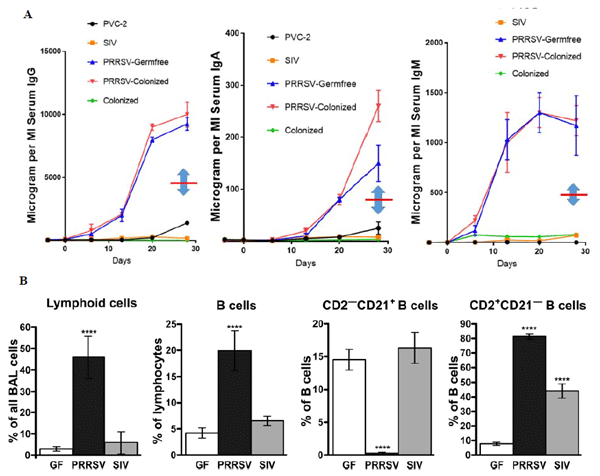
Sinkora M, Butler JE, Lager KM, Potockova H, Sinkorova J. The comparative profile of lymphoid cells and the T and B cell spectratype of germ-free piglets infected with viruses SIV, PRRSV or PCV2. Vet Res, 45:91-1009, 2014.
Sinkora M, Butle, JE. Progress in the use of swine in developmental immunology of B and T lymphocytes. Devel. Comparative Immunology, 58:1-17, 2016.
Sun X-Z, Lager KM, Tobin G, Nara P, Butler JE. Antibody repertoire development in fetal and neonatal piglets. XXIII. Fetal piglets infected with a vaccine strain of PRRS virus display the same immune dysregulation seen in isolator piglets. Vaccine, 30:3646-3652, 2012.
Toman M, Celer V, Kavanova L, Leva L, Frolichova J, Ondrackova P, Kudlackova H, Nechvatolova K, Salat J, Faldyna, M. Dynamics and differences in systemic and local immune responses after vaccination with inactivated and live commercial vaccines and subsequent subclinical infection with PRRS virus. Front. Immunol, doi:3389/fimmun 2019.01689, 2019.
Van Reeth K, Nauwynck H, Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol, S48:325-35,1996.
VanderWall, K, Deen, J. Global trends in infectious disease of swine. Proc Nat’l Acad Sci, 115:11495-11500, 2018
Wang R, Nan Y, Yu Y, Yang Z, Zhang YJ. Variable interference with interferon signal transduction by different strains of porcine reproductive and respiratory syndrome virus. Vet Microbiol, 2013:166:493-503.
Wensvoort G, Terpstra C, Pol JMA, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM , Moonen PLJM , Zetstra T , de Boer EA, Tibben HJ, MF de Jong, P van ‘t Veld, GJR Greenland, JA van Gennep, MTh Voets, Verheijden JHM, Braamskamp J. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q, 1991:13:121-30.
Wilson, MR Emerging viral infections. 26:301-306, 2013,
Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res, 154:48-60, 2010.







