The Historical use of Models in Animal Agriculture
Models of all types have a strong history of application in animal production, where their objectives have typically revolved around optimally feeding and growing livestock. For ~50 years, mathematical models have been assisting researchers, nutritionists and producers with decision making on various aspects of livestock production. A typical trajectory for model development has been (1) the execution of experiments to answer targeted questions and generate data, which undoubtedly (2) raise even more questions and then (3) compels scientists to explore ways to summarize and combine this data, and eventually (4) develop mathematical models to solve the problem of the ever-growing body of data and knowledge being generated and make sense of it. As such, these (predominantly nutritional) models have evolved to mathematically express our cumulative knowledge of how a biological system works, developed in order to understand and manipulate nutrient dynamics in the animal and develop better more efficient and productive feeding programs.
Model Types and Classification
Modelling research scientists often talk about models in terms of their classification, or type, as each has relative strengths and weaknesses and niches for application. For example, models are commonly classified as being static or dynamic, deterministic or stochastic, empirical or mechanistic (Thornley and France, 1984). To delve deeper, a static model is one that makes a prediction at a single time point, whereas a dynamic model is one that considers changes in a biological system over time (typically via a series of integrated differential equations or time step loops). A deterministic model considers the ‘average’ animal and the average animal’s response to an intervention, whereas a stochastic model is probabilistic, and models response variation. An empirical model is a model that studies and describes correlations in the data (e.g. y = mx + b), vs a mechanistic model is a model that describes an underlying causal pathway. To conceptualize the latter, if a cow is represented by level i, then the organs may be represented by level i -1, cells as level i -2 and the herd by level i +1, and a mechanistic model may predict level i outcomes with a mathematical description of level i-1 (always a level lower) attributes(Thornley and France, 1984).
In practice, the lines drawn between model classifications are blurry at best, as, for example, many mechanistic models contain empirical elements, or a deterministic model may have strategic variation/stochastic attributes introduced (e.g. in inputs), but not throughout. Many of the most advanced models in dairy production are mechanistic or quasi-mechanistic (the latter being a hybridization of the empirical and mechanistic approaches, for example the new INRA feeding system for ruminants, Noziere et al., 2018).
Another way models may be classified is whether they are ‘requirement-based’ or ‘responsebased’ models. Requirement based models start with a pre-defined production level, from which they work backwards to determine the nutrient requirements to support that production and then match nutrient supply with requirements. A response model, on the other hand, starts with a description of nutrient supply and the animal, then predicts how the animal will respond to that nutrient supply, and via an optimization routine can find the optimal solution for any objective function (for a nice discussion see Daniel, 2016). While both approaches are useful and applied in practice, a major limitation of the requirement-model approach is that it cannot adapt to the increasing diversity of objectives of dairy production systems (e.g. optimizing vs. maximizing production, feed efficiency, animal health, longevity, etc.). Thus there is a push to increasingly move towards response-based models given their higher level of flexibility.
Ruminant Nutrition Models
Globally, there are numerous groups involved in the development of empirical, quasi-mechanistic and mechanistic models useful for dairy production systems. For an excellent review of the historical evolution of these models, see Tedeschi (2019). In North America, the most commonly used and researched/developed dairy nutrition models include the National Research Council (NRC, 2001), the Cornell Net Carbohydrate and Protein System (CNCPS) (Fox et al., 2004) and Molly (Baldwin, 1995), though other dairy model work exists both within industry and at various academic institutions in North America (for e.g., Dijkstra et al., 1992; Mills et al., 2001; Ellis et al., 2014, as just a few examples). These models have been modified, stepwise, over generations, to account for specific concerns and topics of the era (e.g., performance, efficiency, environmental impact, welfare concerns). The NRC (2001) model is the most commonly used requirement-based model for dairy and could be classified as being quasi-mechanistic, static and deterministic, vs MOLLY (Baldwin, 1995), CNCPS (Fox et al., 2004) and others (Dijkstra et al., 1992; Mills et al., 2001; Ellis et al., 2014) could be classified as being response-models and largely mechanistic, dynamic and deterministic.
Models such as these have been applied over the decades to solve nutritional (Boval et al., 2014; McNamara et al., 2017), whole-farm management (Kebreab et al., 2019) and sustainability (Bannink et al., 2010; Ellis et al., 2011; Gregorini et al., 2013; Van Amburgh et al., 2019) challenges in collaboration with research and extension (many application examples in the literature).
The Role of Mechanistic Models in Dairy Production
Intellectually and academically, mechanistic models (MM) (and to an extent empirical/quasimechanistic models) provide animal scientists the opportunity to explore how a biological system works, extract meaningful information from data (e.g., metabolic fluxes from isotope enrichments (France et al., 1999), and increase our understanding of complex biological systems. They are often used to summarize experimental data to derive meaningful parameters used in other applications, for example, fractional rates of rumen degradation (France et al., 2000) or specific rates of mammary cell proliferation (Dijkstra et al., 1997). In research, MMs are excellent tools for identifying areas where scientific knowledge is lacking, or where a hypothesis on the regulation of a system may be wrong. As such, failure of a MM to simulate reality indicates an area where the system has not been appropriately described, and this could be due to a false assumption, a lack of appropriate data or because the level of aggregation at which the model runs is not appropriate for the research question. When models interact iteratively with animal experimentation, MMs assist movement of the whole field forward by increasing our biological knowledge.
In the field, MMs typically serve as ‘decision-support systems’ or ‘opportunity analysis’ tools, and thus go above and beyond least cost diet formulation. Here we might define ‘decision-support’ as the ability to assist with complex (nutritional, management, etc.) decision making, and ‘opportunity analysis’ as the ability to examine a variety of scenarios for their potential outcomes, with the goal of improving performance, reducing cost, and/or minimizing environmental impact (e.g. Ferguson, 2015). Mechanistic models have been used to solve problems such as: (1) identification of performance limiting factors; (2) determination of the optimal nutrient contents of a feed; (3) evaluation of management factors to optimize performance (Ferguson, 2015); (4) the implementation of strategies to reduce nutrient excretion into the environment (Pomar and Remus, 2019); and (5) forecasting outcomes in scenarios not yet seen in practice (Ferguson, 2015).
Limitations of the MM approach revolve around their manual nature, often extensive input requirements and developer/end-user training requirements. From the end-user perspective, these challenges may mean that MMs may not be user-friendly or approachable enough to guarantee user uptake. Use in the field is typically limited to ‘expert users’ (for example within a nutrition company), as opposed to being directly in the hands of producers. This may, however, be on the verge of changing, with increasing use of sensors, technical training and automated data collection on farm.
The Big Data Wave
The term “big data” has gained considerable attention in recent years, though its definition tends to differ across disciplines (Morota et al., 2018). Common themes in ‘big data’ definitions are: (1) volume: that the volume of data is so large that visual inspection and processing on a conventional computer is limited; (2) data types: may include digital images, on-line and off-line video recordings, environment sensor output, animal biosensor output, sound recordings, other unmanned real-time monitoring systems; and (3) data velocity: the speed with which data are produced and analysed, typically in real-time. In order to gain insight from these large volumes of readily available data, it has become increasingly popular to apply data mining and machine learning (ML) methodologies to cluster data, make predictions or forecast in real-time. Thus, the topics ‘big data’ and ML, though not explicitly tied together, often work hand-in-hand.
The emergence of big data and its associated analytics is visible in scientific referencing platforms such as Scopus, where yearly ‘big data’ references rose from 680 in 2012, to 16,562 in 2018. When combined with the keywords ‘cattle’, ‘pigs’ or ‘poultry’, the first reference to ‘big data’ appears in 2011, but there are only 172 total references from 2011-2018 inclusive, indicating a much slower development rate within animal production systems. Liakos et al. (2018) highlighted that 61% of published agriculture sector papers using ML approaches were from the cropping sector, 19% in livestock production and 10% in each of soil and water science, respectively. There may be several reasons for the slower adoption rate in animal production systems, including the current degree of digitalization, the utility offered, low/unclear value proposition, return-on-investment (ROI), and the challenge of maintaining sensitive technology in corrosive, dusty and dirty environments.
Machine Learning Models
Artificial intelligence (AI) refers to an entire knowledge field focused on the development of computer systems able to perform tasks that normally require human intelligence (e.g. visual perception, speech recognition, decision-making). Machine learning (ML), is actually a subgrouping within AI, focused on the development of algorithms and statistical models used to perform specific tasks, without explicit instructions, relying on patterns and inference within the data. This must already sound familiar, and in fact, several ML approaches are already familiar to us – such as linear and nonlinear regression and principal component analysis. Machine learning represents an umbrella term covering numerous empirical modelling methods, including those most common to us.
A broader categorization of ML models (beyond traditional regression), that fits with the scope of big data would be grouping based on the type of learning used therein (supervised, unsupervised), the nature of the data (continuous, discrete) and the category of problems they solve (classification, regression, clustering, dimensionality reduction). In supervised learning, as it pertains to ‘big data’, ML systems are presented with inputs and corresponding outputs, and the objective is to construct a general rule, or model, which maps the inputs to outputs. By ‘learning’, we refer to the ability for the model to improve predictive performance through an iterative process over time on a defined ‘training dataset’. Compared to the MM approach where evaluation of model performance is generally manual, ML systems automate this step and iterate towards the best model. The performance of the ML model in a specific task is defined by a performance metric such as minimizing the residual error or mean square error, the same tools applied within MM evaluations. The goal is that the ML model will be able to predict, classify or reduce the dimensionality of new data using the experience obtained during the training process.
Conversely, in unsupervised learning, there is no distinction between inputs and outputs, and the goal of the learning is to discover groupings in the data. ‘Clustering’ is a type of unsupervised learning problem aimed to find natural groupings, or clusters, within data. Examples of clustering techniques include k-means (Lloyd, 1982), hierarchical clustering (Johnson, 1967) and the expectation maximisation. Dimensionality reduction, another unsupervised learning method, is the process of reducing the number of variables under consideration by reducing their number to a set of principal variables (components). Principle component analysis (PCA) is a common example of a widely applied dimensionality reduction technique.
Machine learning methods have been shown to help solve multidimensional problems with complex structures in the pharmaceutical industry and medicine, as well as in other fields (LeCun et al., 2015). In this respect, they represent a very powerful data synthesis technique. Similarly, Liakos et al. (2018) found in a review that within the agriculture sector, papers using ML approaches largely focused on disease detection and crop yield prediction. The authors reflected that the high uptake in the cropping sector likely reflects the data intense nature of crop production and the extensive use of imaging (spectral, hyperspectral, near-infrared, etc.). Based on a review of the available literature, currently the application ‘niche’ occupied by ML/big data models in animal production revolve around ‘pattern recognition’ (encompassing classification, clustering) and ‘predictive ability’.
Pattern Recognition
Broadly, ML methodologies demonstrate strength interpreting various types of novel data streams (e.g. audio, video, image) to cluster, classify or predict based on supervised or unsupervised approaches and mapping of patterns within the data. Within animal production, this has most notably been applied to animal monitoring and disease detection.
For example, a series of sensor types, with ML models behind them to interpret and classify the data, have been developed for use in practice to monitor changes in animal behaviour (which may signify a change in health status, injury, heat or energy expenditure level) or are used for animal identification. Numerous publications have shown the ability of sensors to classify animal behaviour (grazing, ruminating, resting, walking, etc.), for example via 3-axis accelerometers and magnetometers (Dutta et al., 2015), optical sensors (Pegorini et al., 2015) or depth video cameras (Matthews et al., 2017), along with ML models to classify the collected data. As continuous human observation of livestock to the extent that a subtle change in behaviour could be observed and early intervention applied is often impractical, the niche for automated monitoring systems to track animal movement and behaviour has formed.
As well, several other examples of how big data and ML has been applied to the task of early disease detection in cattle can be found in the literature. For example, Hyde et al. (2020) used a ML model (random forest algorithm) to automate mastitis detection in dairy herds and specify whether the origin was ‘contagious’ or ‘environmental’. Ebrahimi (2019) and Ebrahimie et al. (2018) have demonstrated the use of ML models to detect clinical and sub-clinical mastitis at the individual cow level based on milk-robot information. In other species, researchers have developed ANN models, which analyse poultry vocalizations in order to detect changes and identify suspected disease status earlier than conventionally possible. For example, Sadeghi et al. (2015) recorded broiler vocalizations in healthy and Clostridium perfringens infected birds. The authors identified five features (clusters of data) using an ANN model, which showed strong separation between healthy and infected birds, and were able to differentiate between healthy and infected birds with an accuracy of 66.6% on day 2 and 100% on day 8 after infection. Early disease detection seems ideally suited to the ML approach, and is likely to see great success and implementation in this realm.
Predictive Abilities
Data driven methodologies have also found a niche in forecasting and predicting (e.g.) numerical outcomes due to their strong fitting abilities and ability to map even minute levels of variation (e.g. within an ANN). As such, within animal production systems they are well situated to forecast performance metrics of economic importance such as body weight (BW), egg production or milk yield. Alonso et al. (2015) used a Support Vector Machine classification model to forecast the BW of individual cattle, provided the past evolution of the herd BW is known. This approach outperformed individual regressions created for each animal in particular when there were only a few BW measures available and when accurate predictions more than 100 days away were required. Pomar and Remus (2019) and Parsons et al. (2007) as well as White et al. (2004) proposed the use of a visual image analysis system to monitor BW in growing pigs from which they could determine appropriate feed allocations, and this was built into their precision feeding system.
Approach Limitations
Limitations of the ML approach include (1) the risk of over-fitting the data, whereby the ML model learns the ‘noise’ in the data and is unable to make adequate predictions on new data, (2) the data volume requirements are high (ML models are ‘data hungry’) in order to avoid biased and skewed predictions, and (3) the lack of transparency behind each prediction. While the MM is a fully ‘white box’ approach – the reasoning behind predictions is fully visible and the logic can be followed, many ML methodologies are ‘black box’, meaning a prediction is produced, which may in fact be a very good prediction, but a causal explanation or rationale for the prediction is absent.
As another perspective on approach limitations, Rutten et al. (2013), as later summarized by Liebe and White (2019), examined published ‘big data’ analytics used for precision management, and classified them as they fell within four categories, being: (1) techniques, (2) data interpretation, (3) integration of information and (4) decision making. They found that most animal agriculture analytics fell into categories (1) and (2) and that integration of these information ‘tools’ into onfarm decision making systems was largely abscent (though we may be on the cusp of this development). Hyde et al., (2020) also commented that despite the large quantity of research into the use of ML technologies to impact (human) clinical management of patients, that examples of translation into an effect on clinical management are seldom found (Clifton et al., 2015). This is an interesting phenomena, which warrants further exploration of the value and barriers to implementation of these new technologies (a few suggestions of which we have provided above), as within animal production systems big data/ML technologies have yet to be widely adopted.
Precision Dairy Nutrition Systems
Precision nutrition is ‘the’ arena where we will see full integration of MM and ML approaches to develop automated knowledge-based nutritional decision-making for dairy. Success in swine (Pomar et al., 2015) and poultry (Zuidhof, 2020) has been demonstrated, and the impact of feeding individuals vs the herd has been explored in dairy (Little et al., 2016; Henriksen et al., 2019). Within such approaches, big data/ML technologies are involved in the monitoring of individual animals (intake, performance, behaviour, health) and MMs are involved in decision making.
The precision feeding approach has developed because feeding the herd as a group typically implies satisfying the requirements of either (1) the average animal or (2) the highest producing animals, resulting in either (1) half the herd receiving less than their requirements or (2) most individuals receiving more nutrients than required. The latter is often the approach, as for most nutrients, underfed individuals may have reduced performance while overfed individuals may still have near optimal performance. Therefore, providing an excess of nutrients ensures that herd performance is not compromised, though it (negatively) results in economic and environmental waste. As an alternative, precision nutrition targets feeding small-group or individually tailored diets, laying the foundation to address key issues in animal production systems, such as (1) reducing feed cost by improving efficiency, (2) improving sustainability and reducing nutrient excretion into the environment, and (3) improving food safety via traceability (Pomar et al., 2015). Precision nutrition is an innovative approach to livestock production based on advanced on-farm monitoring technologies and our cumulative scientific knowledge of animal science, with the objective of optimizing animal production by controlling variability that exists among farm animals and targeting nutrient delivery to the individual (Pomar et al., 2015). However, nutrient ‘requirements’ and performance vary greatly among individual cows within in a herd, in addition to over time as well (through varying physiological phases, stage of lactation, etc.), and thus knowledge of how individuals will respond, based in biology, is a necessity.
To successfully implement such an (automated) precision nutrition system we require (1) precise knowledge of the individuals (identities, milk yield, milk composition, body weight, body condition score, etc. in real-time), (2) a means to forecast next-day performance, and (3) an integrated decision-making tool to guide what should be fed to each individual based on predicted response to nutrient delivery. With the arrival of the ‘big data wave’ in animal production, item (1) is within reason to be collected on farm daily via sensors. To interpret that data and forecast next day performance (2), ML algorithms are ideally suited to this task based on real-time monitoring of (for example) milk production and bodyweight. Mechanistic models logically lie at the heart of item (3), the decision making ‘engine’ within the system – what should be fed to who? In this fully automated system, it is evident that the two seemingly divergent realms of MM and ML occupy related but synergistic ‘niches’, and it is likely that the future of precision nutrition on farm is a hybridization of these approaches, as has been demonstrated in other species (Pomar et al., 2015).
Gargiulo et al. (2018) conducted a survey in Australia, on the uptake and attitude towards precision dairy technologies. First, they found that farmers with more than 500 cows adopted between 2 and 5 times more specific precision technologies (such as automatic cup removers, automatic milk plant wash systems, electronic cow identification systems and herd management software). They also found only minor differences between how large and small farms felt about the future prospects of precision technologies – both were optimistic. Interestingly though, service providers expected a higher adoption of (for example) automatic milking and walk over weighing systems compared to farmers (60.3 vs. 79.4%, respectively). This difference is likely related to the cost and risk related to implementing said technology, variable on-farm performance and unclear monetary and non-monetary benefits (Jago et al., 2013).
The Future of Precision Nutrition
It is likely that the extent to which farms fully digitalize and become ready to utilize these digital technologies will vary globally. As a result, we may never see precision nutrition implemented in all dairy operations, and instead may observe it on a spectrum, such as:
1) No digitalization: Simple empirical models may preside, which can address major issues with easily manipulated equations and minimal input data;
2) Manual data pipeline: A MM model with a custom developed front-end may be most suitable;
3) A small digital pipeline with a limited number (1 or 2) of data streams: May enable automatic input population for application of MM models;
4) A medium digital pipeline: Would mean combining different data sources with management systems delivering real-time data. Some simulation functions of MM may be replaced by real-time variation. The need for heavy-duty front-end development may be reduced.
5) A fully digitized pipeline: Would enable MM and ML models to run a farm, monitoring the status and automatically implement generated recommendations (e.g. Tesla’s AutoPilot, vertical farming).
Within such a categorical analysis of the spectrum, a complete lack of digitalization (1) might suit the use of a simple model by (for example) a 3rd party consultant, based on rough information provided (from memory or estimation) by the farmer. This scenario is often present in underdeveloped countries. The development of a manual data pipeline (2) (e.g. manual measuring and recording of data) allows the development of improved ‘benchmarking’ abilities – and therefore more reliable model predictions from a simple empirical or MM. With a small continuous digital pipeline (3), continuous optimization and real-time benchmarking (as is being achieved now in precision nutrition for swine, e.g. Pomar et al., 2015) becomes possible. It is at this point on the scale of digitalization that things become interesting. From this point forward there also presents a niche for hybridization of the ML and MM approaches, which are described throughout this paper. A medium sized digital pipeline (4) would allow real-time optimization of integral parts of the MM engine, and combine it with digital data. For example, a ration MM-optimized off realtime or ML forecasted intake and BW data, and on the side utilizing a ML model on audio/video data to determine (e.g.) health, activity level or behaviour. A full digital pipeline (5) would allow monitoring and optimization of the entire system. This would allow a MM to be utilized as the basis and optimize/augment with ML learning patterns never envisioned – straight up the ladder of causation (e.g. see Pearl and Mackenzie, 2018). Similar to vertical farming, it would mean the automated management of a farm. To our knowledge, (5) does not yet exist within animal production systems.
As we move towards precision agriculture across species, there may be multiple ways these two modelling approaches will interact. One possible way to achieve this, may be individual parameterization of MMs, via integrating backwards propagation techniques applied within artificial neural networks (ANNs) (for example) into a MM, while placing biological confidence limits around MM parameters permitted to vary between animals to prevent extending beyond biologically sensible values. Knowledge of the biological uncertainty, and setting bounds to limit purely empirical fitting of the model, may be critical to keep the integrity of the MM. Pomar et al. (2015) proposes a real-time closed feedback system to determine nutrient requirements and subsequent feed intakes for individual pigs by combining both MM and ML. A current challenge in applying existing MM response-based models to feeding individuals is that many are developed on pen-fed or data of means of multiple cows, and response of individuals are likely unique (Daniel, 2016). This may require additional attention as we shift our models from the average cow to the individual.
A diagrammatic of how full integration might generate value is presented in Figure 1. Figure 1 summarizes the flow from a problem statement to data to information to knowledge to wisdom (as illustrated in the data-information-knowledge-wisdom pyramid of Ackoff, 1989) and how MM and ML may assist at different steps of that flow. In general, the ML/big data methods may independently only get us so far as the translation of ‘data’ into ‘information’ (perhaps up until knowledge – e.g. disease detection). The MM methods translate ‘knowledge’ into ‘wisdom’, but may lack sufficient ‘information’. Therefore, their integration would seemingly benefit both realms on the path to ‘explainable AI’.
Another approach may be to ‘train’ the biological knowledge from a MM into a ML model, via, for example, training the ML with a latticework of simulations from a MM. Further, as dairy production increasingly experiences demands to simultaneously optimize to multiple criteria (e.g. performance, cost, efficiency and environment), multi-criteria optimization tied to a MM may be required with the biological knowledge of how these factors interact (e.g. see White and Capper, 2014) – utilizing complex parameter optimization algorithms.
The true challenge to widespread adoption of precision feeding in animal production is likely financial and logistical – requiring a substantial investment in facility re-design and technology upgrade. Not until the ROI can be demonstrated, logistical barriers overcome and trust in the system is developed (there is much at stake within animal production if things go wrong), is precision feeding likely to be taken up by the industry as a solution for the future. However, the potential is there to use precision nutrition and automation of the farm as a means to target nutrient delivery, improve performance and herd efficiency, reduce the environmental impact of dairy production and improve overall animal health and wellbeing.
Figure 1. The potential integration of mechanistic models into a decision support system for precision dairy production (Adapted from Ellis et al., 2020).
Summary
Fully automated on-farm precision nutrition is an emerging area within the field of dairy nutrition, targeting the delivery of nutrients to individual animals in order to reduce feed cost, improve feed efficiency and reduce the environmental impact of dairy production. While the concept is not new, the emergence of novel on-farm data streams, allowing producers to follow and monitor the activity, health and productivity of individual animals, is allowing new development in this field. Mechanistic models have served at the heart of nutritional ‘decision-support’ on-farm for decades, and the development of fully automated data collection is opening the door to changes being made on-farm in real-time. The adoption rate of these new technologies is currently relatively slow, and issues such as the current degree of digitalization, access to Wi-Fi on farm, the utility offered, return-on-investment (ROI), and/or the challenge of maintaining sensitive technology in corrosive, dusty and dirty environments will need to be addressed. More than likely, the future of on-farm decision support will be a hybridization of MM and ML technologies, utilizing the respective strengths and augmenting weaknesses of each approach.
Presented at the 2021 Animal Nutrition Conference of Canada. For information on the next edition, click here. 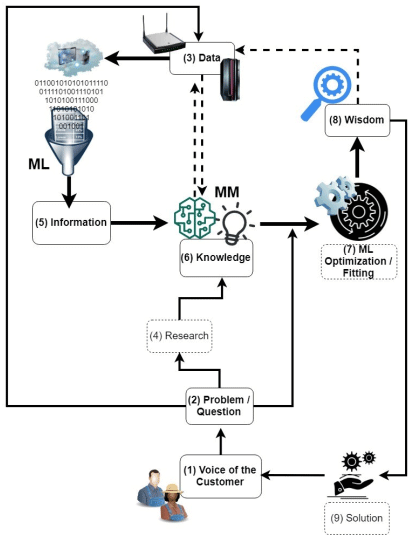







.jpg&w=3840&q=75)