Polymyxin Resistant Bacteria in Australian Poultry
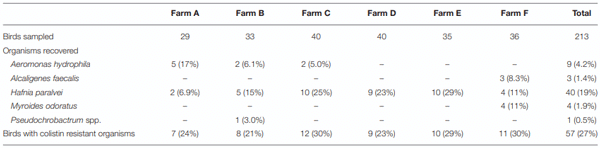
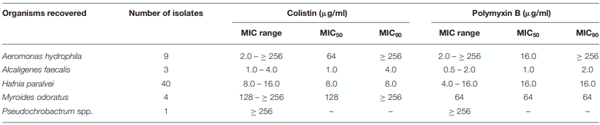
Resistance to last-resort antibiotics is significant public health issue. Antibiotic use in animal husbandry may be a driver of resistance that can subsequently be disseminated via the food chain. This study sought to determine the level of polymyxin resistance in Gram-negative pathogens present in Australian poultry, particularly the presence of mobilizable mechanisms of polymyxin resistance. Cloacal swabs from 213 birds were taken in a point prevalence survey from six different farms at a Victorian chicken processing facility. Colistin resistant organisms were recovered by direct plating on CHROMagar COL-APSE media. Bacterial isolates were identified and analyzed by MALDI-TOF, biochemical and genotypic assays. The 213 specimens yielded 57 (26.8%) colistin-resistant Gram-negative organisms, all of which have been previously described as exhibiting intrinsic resistance to polymyxin antibiotics. The most frequent organism was identified as Hafnia paralvei (40/57; 70%). Other colistin-resistant organisms included Aeromonas hydrophila (16%), Myroides odoratus (7%), Alcaligenes faecalis (5%), and Pseudochrobactrum spp. (2%). No mobile colistin resistance (mcr) genes were detected, although the arnA gene was identified in two A. hydrophila isolates and may mediate colistin resistance in these isolates. Intrinsic polymyxin-resistant organisms are widely distributed in the food chain, with over a quarter of the birds tested yielding a polymyxin-resistant organism. However, strains containing mcr genes remain rare in Australian poultry.
Keywords: Aeromonas, polymyxin-resistance, colistin, poultry, Hafnia paralvei, antibiotics.
INTRODUCTION
MATERIALS AND METHODS
Specimen Collection
Bacterial Identification
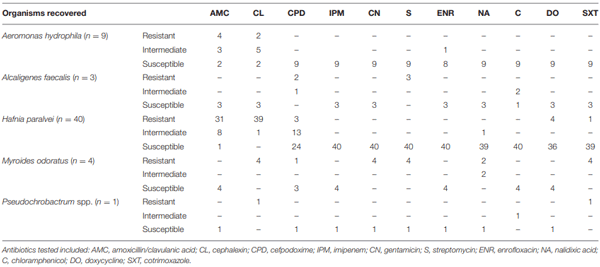
Antimicrobial Susceptibility Testing and Determination of Minimal Inhibitory Concentrations (MICs)
Detection of MCR-like Genes
DNA Sequencing
RESULTS
DISCUSSION
DATA AVAILABILITY STATEMENT
ETHICS STATEMENT
AUTHOR CONTRIBUTIONS
ACKNOWLEDGMENTS
Abbott, S. L., Moler, S., Green, N., Tran, R. K., Wainwright, K., and Janda, J. M. (2011). Clinical and laboratory diagnostic characteristics and cytotoxigenic potential of Hafnia alvei and Hafnia paralvei strains. J. Clin. Microbiol. 49, 3122–3126. doi: 10.1128/JCM.00866-11
Abdul Momin, M. H. F., Bean, D. C., Hendriksen, R. S., Haenni, M., Phee, L. M., and Wareham, D. W. (2017). CHROMagar COL-APSE: a selective bacterial culture medium for the isolation and differentiation of colistin-resistant gram-negative pathogens. J. Med. Microbiol. 66, 1554–1561. doi: 10.1099/jmm.0.000602
Animal Health Australia (2017). Animal Health in Australia 2016. Canberra, ACT: Animal Health Australia.
Apostolakos, I., and Piccirillo, A. (2018). A review on the current situation and challenges of colistin resistance in poultry production. Avian Pathol. 47, 546–558. doi: 10.1080/03079457.2018.1524573
Aravena-Roman, M., Inglis, T. J., Henderson, B., Riley, T. V., and Chang, B. J. (2012). Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob. Agents Chemother. 56, 1110–1112. doi: 10.1128/AAC.05387-11
Arcilla, M. S., Van Hattem, J. M., Matamoros, S., Melles, D. C., Penders, J., de Jong, M. D., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16, 147–149. doi: 10.1016/S1473-3099(15)00541-1
Australian Chicken Meat Federation (2018). Position Statement: Responsible use of Antibiotics in the Australian Chicken Meat Industry. Sydney, NSW: Australian Chicken Meat Federation.
Australian Pesticides and Veterinary Medicines Authority (2014). Quantity of Antimicrobial Products Sold for Veterinary use in Australia: July 2005 to June 2010. Kingston, NY: Australian Pesticides and Veterinary Medicines Authority.
Breazeale, S. D., Ribeiro, A. A., Mcclerren, A. L., and Raetz, C. R. (2005). A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 280, 14154–14167. doi: 10.1074/jbc.M414265200
Buess, S., Nuesch-Inderbinen, M., Stephan, R., and Zurfluh, K. (2017). Assessment of animals as a reservoir for colistin resistance: no MCR-1/MCR-2-producing Enterobacteriaceae detected in Swiss livestock. J. Glob. Antimicrob. Resist. 8, 33–34. doi: 10.1016/j.jgar.2016.11.001
Chen, L., Zhang, J., Wang, J., Butaye, P., Kelly, P., Li, M., et al. (2018). Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS ONE. 13:e0193957. doi: 10.1371/journal.pone.0193957
Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Eighth Informational Supplement CLSI M100-S28. Wayne, PA: CLSI.
Doulgeraki, A. I., Paramithiotis, S., and Nychas, G.-J. E. (2011). Characterization of the Enterobacteriaceae community that developed during storage of minced beef under aerobic or modified atmosphere packaging conditions. Int. J. Food Microbiol. 145, 77–83. doi: 10.1016/j.ijfoodmicro.2010.11.030
Ellem, J. A., Ginn, A. N., Chen, S. C., Ferguson, J., Partridge, S. R., and Iredell, J. R. (2017). Locally acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013. Emerg. Infect Dis. 23, 1160–1163. doi: 10.3201/eid2307.161638
Fosse, T., Giraud-Morin, C., and Madinier, I. (2003). Induced colistin resistance as an identifying marker for Aeromonas phenospecies groups. Lett. Appl. Microbiol. 36, 25–29. doi: 10.1046/j.1472-765X.2003.01257.x
Gatzeva-Topalova, P. Z., May, A. P., and Sousa, M. C. (2005). Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure 13, 929–942. doi: 10.1016/j.str.2005.03.018
Gunzer, F., Rudolph, W. W., Bunk, B., Schober, I., Peters, S., Müller, T., et al. (2018). Whole-genome sequencing of a large collection of Myroides odoratimimus and Myroides odoratus isolates and antimicrobial susceptibility studies. Emerg. Microbes Infect. 7:61. doi: 10.1038/s41426-018-0061-x
Höll, L., Behr, J., and Vogel, R. F. (2016). Identification and growth dynamics of meat spoilage microorganisms in modified atmosphere packaged poultry meat by MALDI-TOF MS. Food Microbiol. 60, 84–91. doi: 10.1016/j.fm.2016.07.003
Holmes, B., Snell, J. J., and Lapage, S. P. (1979). Flavobacterium odoratum: a species resistant to a wide range of antimicrobial agents. J. Clin. Pathol. 32, 73–77. doi: 10.1136/jcp.32.1.73
Huys, G., Cnockaert, M., Abbott, S. L., Janda, J. M., and Vandamme, P. (2010). Hafnia paralvei sp. nov., formerly known as Hafnia alvei hybridization group 2. Int. J. Syst. Evol. Microbiol. 60, 1725–1728. doi: 10.1099/ijs.0.018606-0
Jacquier, H., Le Monnier, A., Carbonnelle, E., Corvec, S., Illiaquer, M., Bille, E., et al. (2012). In vitro antimicrobial activity of “last-resort” antibiotics against unusual nonfermenting Gram-negative bacilli clinical isolates. Microb. Drug Resist. 18, 396–401. doi: 10.1089/mdr.2011.0195
Jayol, A., Saly, M., Nordmann, P., Menard, A., Poirel, L., and Dubois, V. (2017). Hafnia, an enterobacterial genus naturally resistant to colistin revealed by three susceptibility testing methods. J. Antimicrob. Chemother. 72, 2507–2511. doi: 10.1093/jac/dkx154
Kämpfer, P., Rosselló-Mora, R., Scholz, H. C., Welinder-Olsson, C., Falsen, E., and Busse, H.-J. (2006). Description of Pseudochrobactrum gen. nov., with the two species Pseudochrobactrum asaccharolyticum sp. nov. and Pseudochrobactrum saccharolyticum sp. nov. Int. J. Syst. Evol. Microbiol. 56, 1823–1829. doi: 10.1099/ijs.0.64256-0
Kaur, M., Shang, H., Tamplin, M., Ross, T., and Bowman, J. P. (2017). Culturedependent and culture-independent assessment of spoilage community growth on VP lamb meat from packaging to past end of shelf-life. Food Microbiol. 68, 71–80. doi: 10.1016/j.fm.2017.06.015
Ling, Z., Yin, W., Li, H., Zhang, Q., Wang, X., Wang, Z., et al. (2017). Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 61:e01272-17. doi: 10.1128/AAC.01272-17
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Ma, S., Sun, C., Hulth, A., Li, J., Nilsson, L. E., Zhou, Y., et al. (2018). Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 73, 1777–1780. doi: 10.1093/jac/dky110
Monte, D. F., Mem, A., Fernandes, M. R., Cerdeira, L., Esposito, F., Galvão, J. A., et al. (2017). Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 61:e02718–16. doi: 10.1128/AAC.02718-16
Mukerji, S., Stegger, M., Truswell, A. V., Laird, T., Jordan, D., Abraham, R. J., et al. (2019). Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J. Antimicrob. Chemother. 74, 2566–2574. doi: 10.1093/jac/dkz242
Okada, S., and Gordon, D. M. (2003). Genetic and ecological structure of Hafnia alvei in Australia. Syst. Appl. Microbiol. 26, 585–594. doi: 10.1078/0723202037708 65891
Olaitan, A. O., Morand, S., and Rolain, J.-M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Säde, E., Murros, A., and Björkroth, J. (2013). Predominant enterobacteria on modified-atmosphere packaged meat and poultry. Food Microbiol. 34, 252–258. doi: 10.1016/j.fm.2012.10.007
Shen, Y., Xu, C., Sun, Q., Schwarz, S., Ou, Y., Yang, L., et al. (2018). Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 62:e00404- 18. doi: 10.1128/AAC.00404-18
Teale, C. J. (2002). Antimicrobial resistance and the food chain. J. Appl. Microbiol. 92, 85S−89S. doi: 10.1046/j.1365-2672.92.5s1.20.x
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bact. 173, 697–703. doi: 10.1128/JB.173.2.697-703.1991
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmidmediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17
Zhang, Q. Q., Han, Y. Q., Cao, J. X., Xu, X. L., Zhou, G. H., and Zhang, W. Y. (2012). The spoilage of air-packaged broiler meat during storage at normal and fluctuating storage temperatures. Poult. Sci. 91, 208–214. doi: 10.3382/ps.2011-01519









.jpg&w=3840&q=75)