Background:
FAdVs are widely distributed, and some species are associated with important poultry diseases, representing current threats of serious economic losses for the aviculture industry. Some of the diseases include the Inclusion body hepatitis (IBH) related with FAdV-C and D infections, hepatitis-hydropericardium syndrome (HHS) related with FAdV-C infections [1,2], gizzard erosion (GE) related with FAdV-A infections [3-6] among others. The family Adenoviridae is divided into five genera: Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus and Ichtadenovirus (Harrach 2011). Currently, Fowl adenovirus (FAdV) is classified into five different species (FAdV-A to FadV-E) on based upon the DNA restriction patterns generated by endonucleases BamHI and HindIII [7]. Recently, at least 12 genotypes within the five species were revealed by sequence analysis of the hexon loop 1 (L1) gene region [8].
Proteins are the essential immune-target structures, which in the MHC class I/II (MHC-I/II) pathway are processed to 8- 11/15-18 mer peptides. In this way, peptides that bind to MHC molecules are presented and potentially recognized by cytotoxic T cells (CD8/CD4), which can lead to an immune response [9]. The most selective step in this antigen presentation is the peptide binding to MHC [10]. Each MHC molecule has a potentially unique binding affinity motif [11] and the characterization of this motif for each MHC molecule prevalent in a given population is a central aspect of rational T cell epitope discovery. Due to the immense MHC polymorphism [12,13], an exhaustive characterization of all MHC molecules is a high cost-intensive effort, and as of today in spite of significant advances in high-throughput immune assays only a little more than 100 MHC molecules, including 25 non-human molecules have been experimentally characterized at a detail allowing to describe their binding specificity (IEDB date 2012).
To face this problem, several in silico prediction methods have been developed in the last decades [14-18]. Of these methods, NetMHCpan is the current in state-of the art method for predicting binding affinity of peptides to any MHC-I/II molecule with a know protein sequence [19]. In this study we sequence the complete genome of a FAdV-C from Peruvian wild strains, and perform an immunoinformatics analysis to predict immunogenic MHC-I and MHC-II T-cell linear epitopes for immunodiagnostics and potential vaccine.
Methodology:
FAdV isolation and DNA preparation
Field samples from a local chicken farm let us isolate a FAdV strain. Inclusion body hepatitis (IBH) was confirmed by clinical symptoms and by real-time PCR [20]. Liver from each individual was blended with phosphate buffered saline using individual sterile blenders. Then, for virus isolation, IBH PCR positive homogenized livers were ultracentrifuged by CsCl gradient as follows, cellular lysis was reached given tissues three freeze-thaw cycles (-80°C to 37°C). Cellular debris was eliminated through centrifugation of samples to 5,500 g for 30 minutes at 4°C. Five milliliters of supernatant were added into CsCl gradient (δ= 1.25 g/mL and δ= 1.4 g/mL) and ultracentrifuged in a SW28 rotor (Beckman Coulter, Indianapolis, USA) at 35000 RPM for 1 hour at 15°C in a Optima L-100K (Beckman Coulter). Capsids and complete viral particles were clearly identified. An aliquot of complete virus was used for DNA isolation as mentioned before and quantitation by realtime PCR, [20]. For DNA extraction, an aliquot of 200 µl was used to DNA isolation by DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following manufacturer instructions.
FAdV genome sequencing, assembly and annotation
For the genome sequencing we obtained 2 mg of the Phi29- amplified viral DNA, and used for the construction of the library, which was sequenced using the 454-Roche GS-FLX Titanium System (454 Life Sciences, Roche). The reads obtained from the sequencing were assembled de-novo using GS De Novo Assembler 2.7. Additionally, we assembled the reads using as reference the FAdV-C serotype 4 strain ON1 (Accesion Code: GU188428) in the software GS Reference Mapper. Genome annotation and manual revision were performed using the software CLC Main Workbench 6.8.3. All the possible ORFs were predicted. We used as criteria to validate an ORF, the fact of encoding peptides longer than 30 aa, thus generating putative proteins. The amino acid sequences of all the putative proteins were compared to the available protein sequences in GenBank using the Basic Local Alignment Search Tool (BLAST). The presence of homologous genes in other adenoviruses was used as evidence to support our potential genes.
Sequencing of the Cobb chicken MHC-I and MHC-II coding genes
We collected twelve breeding commercial Cobb chickens. All animals were sampled from farms located in Chincha, an important city of chicken breeding located in the mid-southern coast of Peru. Spleen samples were taken from each selected chicken described above. The tissue was preserved in RNALater (Ambion) and stored at -80°C until further processing. Spleen RNA was purified by homogenizing 20 mg of excised tissue in 600 µL of RLT buffer (Qiagen). The sample was mechanically disrupted with the TissueLyser system (Qiagen), then proceeding (the RNA extraction) according to the manufacture’s protocol (RNeasy mini kit; Qiagen) and resuspended in 50 µL of DEPC-treated water. Exceeding DNA was eliminated by Turbo DNase (Ambion) treatment.
cDNA synthesis was performed according to the manufacturer’s instructions (Transcriptor First Strand cDNA Synthesis Kit, Roche); second, to amplify the entire ORF, cDNA was used as template in a conventional PCR, carried out by a High Fidelity polymerase and PCRx Enhancer (Invitrogen). Primers used for alpha subunit of MHC class I were: (Forward) 5’-ATGGGCTCGTGCGGGG-3’ and (Reverse) 5’- CAGATGGAGGGGTTGCTC-3’; and for the beta subunit (βmicroglobulin) were: (Forward) 5’-AATGGGGAAGGCGGCG3’ and (Reverse) 5’-GGATCCCACTTGTAGACCT-3’. Primers used for alpha subunit of MHC class II were: (Forward) 5’- ATGGCGGTGCTGAGCG-3’ and (Reverse) 5’- CAGAGCAGCCCCGGTT-3’; and for the beta subunit: (Forward) 5’- AATGGGGAGCGGGCGCGT-3’ and (Reverse) 5’- TTCAGCATCCCTGGAGCGGC-3’. PCR products were cloned into pGEM-T Easy plasmid (Promega). DNA was sequenced using 454 GS Junior sequencer (Roche). Each amplicon of MHC subunits were sequenced using a different multiplexer identifier (MID). Unlike MHC-Iα and MHC-II subunits, the MHC-Iβ amplicon is small (350 bp length), so this allowed us to discriminate between different subunits; therefore there was no need to clone it because the entire amplicon could be sequenced in one read.
The sequencing experiment produced .sff files containing the information of the reads and the quality by each position. These reads were processed in the software GS Amplicon Variant Analyzer 2.7 to obtain contigs, using as reference the reported sequences of MHC-I alpha subunit (HQ435266.1, EF554701.1, HQ435266.1), MHC-I beta subunit (AB162661.1, EF554713.1, AB162661.1), MHC-II alpha subunit (EF554701.1) and MHC-II beta subunit (EF554713.1). The variations in the sequences were revised in detail in the software GS Amplicon Variant Analyzer 2.7 and CLC Main Workbench 6.8.3, and the most frequent haplotype for each subunit were determined.
Selection of human MHC-I/II alleles that best substitute its chicken homologues
We used the NetMHCcons and NetMHCIIpan immunoinformatics software to predict potential epitopes with high affinity to MHC class I and II, respectively. The MHC alleles available in these tools belong mainly to humans, macaques, mice, and pigs. Because there is no data of alleles of chicken MHCs, we selected “the best human substitute alleles” for the chicken MHC. To find these substitute alleles, all the sequences of the human MHC-I and II alleles available in NetMHCpan and NetMHCIIpan were compared with the most frequent haplotypes of the chicken MHC alleles. After multiple alignments, we selected the human MHC alleles that showed an identity greater than 70%. The selected human MHC alleles and the most frequent haplotypes of the chicken MHC alleles were compared at their peptides “predictive level”, using as criteria, the similarity of the predicted affinities with a random set of 100,000 peptides. This was performed using the software MHCcluster [21].

Prediction of immunogenic MHC-I and MHC-II FAdV peptides Prediction of peptide affinity to MHC-I and MHC-II
We explored the FAdV genomes looking for potential immunogenic peptides. The FAdV proteins were analyzed with NetMHCcons v1.1 (for MHC-I) [18] and NetMHCIIpan v2.2 (for MHC-II) [22], for the prediction of MHC-peptide binding affinity. For MHC-I peptides, NetMHCcons cuts the proteins in all the 9 amino acids length possible peptides. These peptides and “the best human sustitutes” MHC-I alleles were used to predict the MHC-peptide binding affinity. Similarly, the NetMHCIIpan software cuts the proteins in all the 15 amino acids length possible peptides and uses “the best human sustitutes” MHC-II alleles to predict the binding affinity. The predictions were made for all the human-substitute alleles. These predicted binding affinities were estimated as IC50 concentration (in nM). Additionally, these software provide a pre-defined cut-off for the categorization of the peptides according to the predicted affinity: “SB” or Strong Binder (for IC50 ≤ 2 nM) and “WB” or Weak Binder (for 2 nM < IC50 ≤ 50 nM). Higher values of IC50 are considered as evidence of lack of binding.
Filtering of promiscuity, conservation, host proteome, and surface accessibility
Promiscuity: Promiscuous peptides are defined as those recognized by multiple MHC alleles. We determined the peptides with high affinity to the several MHC-I optimal human alleles selected. Only the peptides predicted as strong binders for all the MHC-I alleles were selected. The same procedure was conducted for MHC-II alleles; Conservation: The peptides obtained after the promiscuity filter were aligned with all the proteins of FAdV available in GenBank. Only the strictly conserved epitopes were selected; Host proteome: The peptides filtered at this point were aligned with the chicken proteome, to avoid selecting epitopes present in the host. Surface accessibility: The localization of the selected peptides in their corresponding native proteins was determined to verify if the peptide is exposed to the surface and thus could be recognized by an antibody. This step was performed only for MHC-II peptides.
Design of the FAdV multiepitopic protein and its encoding gene
The selected MHC-I and MHC-II peptides were concatenated linearly in two synthetic designed protein constructs, called “multi-epitopic proteins”, one for the MHC-I peptides and the other for the MHC-II peptides. These proteins were designed avoiding the appearance of strong binder peptides in the junctions between two consecutive peptides. To produce the recombinant multiepitopic proteins, ad-hoc designed genes encoding them were synthesized, including and optimization of the codon usage for E. coli K12, where these genes were expressed. The synthetic genes were cloned in pET28a-c plasmid, using the NcoI and XhoI restriction sites, including a six-His-Tag at the 3’ end for affinity chromatography purification. These genes were synthesized by the company Genemed Synthesis Inc. (http://www.genemedsyn.com/).
Expression and purification of the FAdV MHC-I and MHC-I multiepitopic proteins
The plasmids with the synthetic genes were inserted in E. coli competent cells (Novablue) by electroporation. The correctly transformed colonies were selected in petri dishes with LuriaBertani agar culture supplemented with 30 µg/mL of kanamycin. To produce each MHC-I and MHC-II multiepitopic protein, the transformed E. coli cells were induced by the addition of isopropil-1-tio-ß-D-galactopyranoside (IPTG) 1mM. The expressed protein was isolated from the bacteria by thermal shock, sonication and centrifugation. The thermal shock consisted in incubation at -70°C during 10 min, then 37°C for 10 min, and this procedure was repeated three times. The sonication was performed using 15,000 g at 4°C during 20 min. The multi-epitopic proteins will be obtained in the no-soluble phase of the inclusion bodies. This phase was solubilized adding urea 8 M. The solubilized fraction was purified by affinity chromatography using His-trap columns. To remove any E. coli protein of low-med affinity to the His-trap columns, solutions with discrete incremental concentrations of imidazole were eluted in the column. The solutions were used in a progressively increasing imidazole concentration, until the multiepitopic protein was eluted. The presence of the multiepitopic protein was verified in a SDS-PAGE electrophoresis revealed with Coomasie blue. The resulting protein was aliquoted in siliconated tubes of low proteinretention in volumes of 500 µl and stored at -70°C.
Immunization of SPF chickens with the FAdV MHC-II multiepitopic protein and antibody detection
Forty-eight, 3-week-old SPF chickens were selected and separated into four groups. Twelve were immunized subcutaneously once with 25 µg of multiepitopic protein with QuilA adjuvant at day 1, while 24 chickens were immunized with the same volume of adjuvant, and the 12 remaining chickens remained with no immunization. The pre-immune sera were collected on the first day. Post-immune sera was collected at the second, third and fourth week for detection of antibodies
Serum antibodies to multiepitopic proteins were detected using a standard ELISA assay. Briefly, after washing the fixed plates with the multiepitopic proteins, 50 ml of the test serum diluted 1:10 in phosphate buffer pH 7.2 was added. After incubation at 37°C for 1 h, 100 µL horseradish peroxidase conjugate, diluted 1:1000, was added to each well and plates were further incubated at 37° C for 1 h. After two washing steps, 100 mL of TMB substrate was added and incubated at room temperature for 10 min. The reaction was stopped by adding 100 ml of H2SO4 2M. Absorbance was measured at 490 nm with a micro ELISA reader (Bio-Rad).
Results:
Genome sequence
The full genome of FAdV-C was found to be 45,673 bp with a G+C content of 54.63%, and very similar to the other FAdV strains, comprising a total of 44 coding genes.
Human MHC-I and MHC-II best chicken-substitute-alleles
We found nine human MHC-I alleles highly similar to the chicken MHC-I. These sequences were used in the MHCcluster, where by means of the heatmap were determined three human alleles as best substitutes. These alleles were HLA*B 40:06, HLA*B 41:04 and HLA*B 41:03 (Figure 1A). In the same way, 56 MHC-II human sequences similar to the alpha chain of the chicken MHC II were selected and analyzed by the heatmap of the MHCcluster. DRB1 alleles 1482, DRB1:1366, DRB1:1310 and DRB1:1445 were found as the best human substitute alleles of the chicken MHC class II (Figure 1B).
FAdV MHC-I and MHC-II peptides predicted as immunogenic in chicken
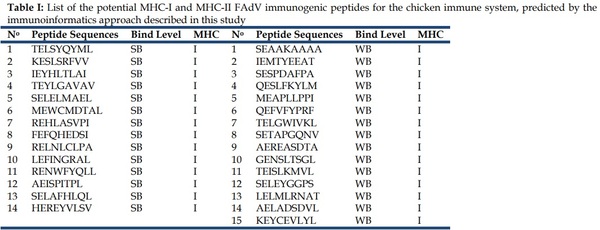
After all the analyses and filters described before, a list of 29 MHC-I peptides and 9 MCH-II peptides from FAdV were selected as potentially immunogenic in chicken (Table I).
Immunogenicity of FAdV MHC-II multiepitopic protein in chickens
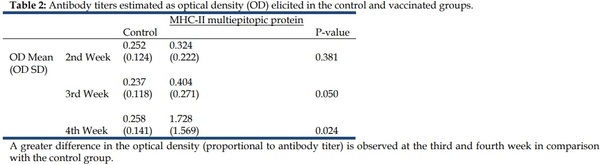
Antibody stimulation experiments were conducted using the multiepitopic MHC-II protein. Chickens were immunized and anti- MHC-II-multiepitopic-protein antibodies were measured. Compared to the control group, a significant increase in the anti MHC-II-multiepitopic protein antibodies was observed in the immunized group four weeks after the first immunization. After the booster, the anti-MHC-II-multiepitopic protein antibodies level increased notably and remained at a high level throughout the duration of the experiment. This confirms that the immune system of the chicken was activated by the immunization with the MHC-II multiepitopic protein (Table 2).
Discussion:
This study shows for the first time an immunoinformatics analysis of the complete FAdV genome to determine potential MHC-I and MHC-II immunogenic peptides for the chicken immune system. The genome sequence of the Peruvian FAdV strain confirmed its global conservation while clustering within other strains. Linear peptides with predicted high affinity to the Cobb chicken MHC-I and MHC-II complexes were found to also meet criteria of specie specificity, MHC allele promiscuity, and conservation through the FAdV genome sequences analyzed here. This is an important evidence for these peptides to be potentially immunogenic [23]. In the case of MHC-II linear T-cell predicted peptides, these were filtered for surface accessibility. MHC-II linear epitopes that are exposed to the surface are potential candidates to elicit a neutralizing antibody response, which is important during viral infections, in order to control the circulating viruses [24,25].
This article was originally published in Bioinformation 11(10): 460-465 (2015). This is an Open Access article distributed under the terms of the Creative Commons Attribution License.