Main results and current investigations on sweetpotato whitefly chemosensory proteins
Published: September 1, 2021
By: Liu Guoxia, Xuan Ning, Guo Xia, HongMei Ma, Jean-François Picimbon / Biotechnology Research Center, Shandong Academy of Agricultural Sciences, Jinan 250100, P.R. China.
Screening of EST-database from the whitefly B. tabaci allowed identification of three BtabCSP clones (BtabCSP-1, BtabCSP2 and BtabCSP3; Liu et al., 2016a,b). In the course of our studies on whitefly CSPs, we have shown that BtabCSPs and in particular BtabCSP2 contain specific genomic DNA sequences (Q260, BaA, CWF-1, BaA, OPT12 and SCAR markers) that can become extremely useful for biotyping. BtabCSPs represent a set of functional “nuclear” genes that are as important as evolutionary markers than internal transcribed spacer (ITS) and mitochondrial cytochrome oxidase I DNA (CoxI) –Figure 1. Accordingly, we attempt to develop a new assay or kit that will employ fluorescent CSP DNA-probe to characterize the biotype in the context of whitefly control –Figure 2. So far, we have successfully developed in China another method (BtabCSP-CAPS) particularly useful for Bemisia whitefly biotyping. BtabCSP-cleaved amplified polymorphic sequence (CAPS) clearly differentiates B and Q biotypes among various B. tabaci field collections from Shandong Province –Figure 3 (Liu et al., 2016b). Fluorescent Q260 and BtabCSP CAPS are profound results for whitefly research and agriculture. Accurate biotype identification is condicio sine qua non for a true efficient insect pest control method.
In our study of genetics and genomics underlying the CSP gene family in whiteflies, we have also found a very intriguing result in relation with epigenetic regulation of gene expression. As found in Bombyx, we have observed that CSPs in B. tabaci are subject to specific RNA mutations and editing mechanisms (Xuan et al. , 2014, 2016, 2019; Liu et al. , 2014, 2016a,b, 2017, 2019, 2020). Very interestingly, investigating RNA variance on BtabCSP, we found significant differences between these two distinct genetically characterized biotypes, B and Q, suggesting that RNA editing in CSP gene family may be linked to multiple insecticide resistance.
Particularly high levels of RNA editing in flour beetle Tribolium castaneum CSPs are found in the gut, perhaps in intimate association with gut microbiota and metabolic process (Liu et al., 2017). Similarly to Tribolium strains, the females from the biotype-Q are known to have a particularly strong insecticide resistance capacity. Therefore, in order to study the function of CSP in particular in relation with gut, metabolism and insecticide resistance, we measured BtabCSP gene expression levels following different insecticide treatments (time and dosage). In moths, we show that most of all BmorCSP genes are up-regulated in many various tissues after avermectin insecticide exposure, including in particular the gut and fat body (Xuan et al. 2015, Insect Science Award, Most Cited Article of 2017). In Bemisia, CSP-1 expression was significantly up-regulated after treating whiteflies with 50 mM neocotinoid thiamethoxam. We show that CSP-1 does not bind insecticide but linoleic acid, strongly suggesting that CSP mediates biotype-specific expression of resistance to insecticide not by a direct interaction with toxic molecules, but by mediating the transport of long fatty acid chains, i.e. fuels for immune response. This may contribute to a very general adaptive mechanism of cells and organisms to environment - Figure 4 (Liu et al. , 2014, 2016a,b, 2017, 2020). In addition, we show that other CSPs in whiteflies do not bind lipids, but rather small insecticide toxic molecules of plant oil origin such as α-pentyl-cinnamaldehyde, demonstrating that CSPs are involved at many different levels of insect defense, i.e. in many different forms of the immune response of the insect exposed to chemical stress (Xuan et al., 2015; Liu et al., 2016b, PLOS ONE, 10% most cited) –Figures 4&5.
Biochemical, molecular and structural analyses are underway to compare tissue-expression levels of CSP variant in between sexes and biotypes. Cloning ADAR and/or other editing enzymes coupled to functional assay is under investigation. Consequently, functional assays are adopted in an approach whereby each protein under investigation is analyzed while considering its tissue distribution profile. The protein partners of CSPs reside in cytochromes P450, degrading enzymes, membrane receptors and/or other binding protein families (OBPs?) for the degradation of environmental xenobiotics and/or insecticide signaling, which should be revealed by future protein-protein interaction studies.
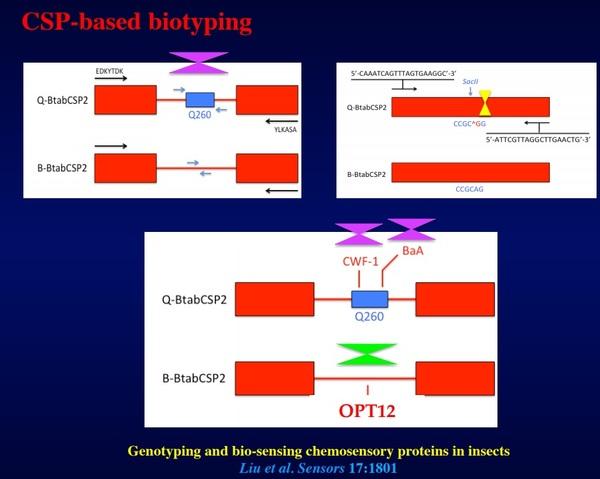
Figure 1. BtabCSP2-based method of biotype recognition in the sweetpotato whitefly, B. tabaci. The triangles in purple show BtabCSP2-specific genomic DNA markers in Q: Q260, CWF-1 and BaA. Recognition of Q is based on two-steps PCR: 1) amplification of BtabCSP2-genomic DNA (black arrows), 2) re-amplification using primers specifically tuned to Q260 sequence (blue arrows). No product is found for B (lacks Q260). The same approach can be used for re-amplification of CWF-1 or BaA marker on BtabCSP2. For recognition of B, the approach adopts use of primers tuned to SCAR marker OPT12 (#EU660887, inverted position, triangle in green). Recognition of Q can also be done using SacII restriction enzyme that recognizes CCGC^GG sites and cuts best at 37°C (see triangle in yellow). Cutting will not occur in B biotype (^G>A mutation site, “no cleavage”). The final result will contain different products using PCR after SacII
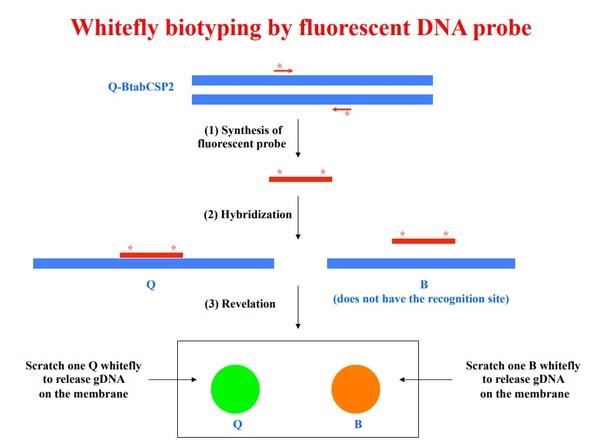
Figure 2. Fluorescent assay for recognition of BtabCSP2 in Q biotype. This “intron” approach takes the synthesis of specific DNA probes by PCR and specific amplification of Q260 (*). *Q260 probe is exposed to gDNA, only gDNA from Q biotype will hybridize with the probe tuned to Q260-intron (labelling with green fluorescence)
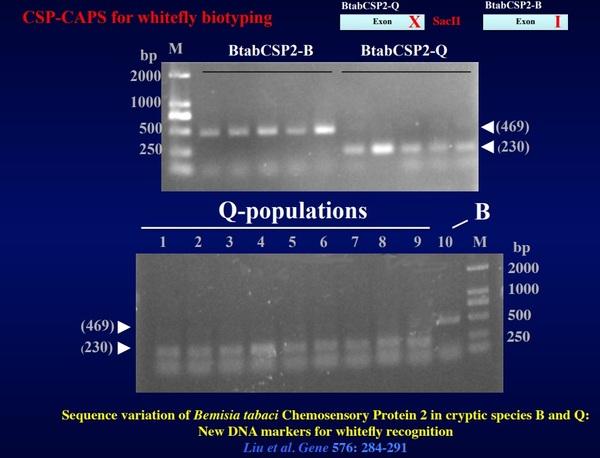
Figure 3. BtabCSP-cleaved amplified polymorphic sequences (CAPS) for B. tabaci biotyping. BtabCSP-CAPS has been developed to differentiate B and Q based on the sequence variation in “exon” of BtabCSP2 (biotype-specific mutation). Q: SacII site “CCGCGG”, B: non SacII site “CCGCAG” (uncut). Different gel agarose electrophoresis profiles between B and Q of BtabCSP2-PCR after Sac II digestion (BtabCSP2-CAPS). B: 469 bps, Q: 230 bps. Applying BtabCSP-CAPS to whitefly collections from cotton in Dezhou and Qingdao shows high prevalence of Q-biotype in Shandong Province. This information is crucial to design most appropriate Bemisia control method before to spray insecticides in the Province
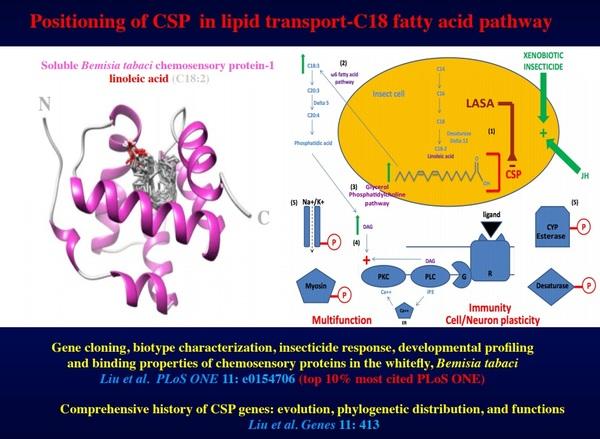
Figure 4. Knock out of BtabCSP1 for inhibition of C18 pathway and insect immune response. Three-dimensional structure of Bemisia tabaci chemosensory protein-1 bound to linoleic acid (in grey). Positioning of whitefly CSP-1 in linoleic acid transport, omega6 fatty acid pathway, glycerol phosphatidylcholine pathway, synthesis of diacylglycerol (DAG), activation of protein kinase C and phosphorylation of proteins involved in many different functions in the insect (neural transmission, locomotion, degradation of toxins and insecticides, insect defense, immunity, cell growth and neurogenesis): ion channels, myosin, cytochrome P450, esterase, desaturase and ligand receptors, among many others. Using LASA (Linoleic Acid Suppressing Agent) may block the functional binding sites of CSP1 thereby interfering with transport of linoleic transport and all cellular mechanisms dependent of C18-lipid pathways. The insect becomes particularly sensitive to toxins, xenobiotics, insecticides and/or insect growth regulators
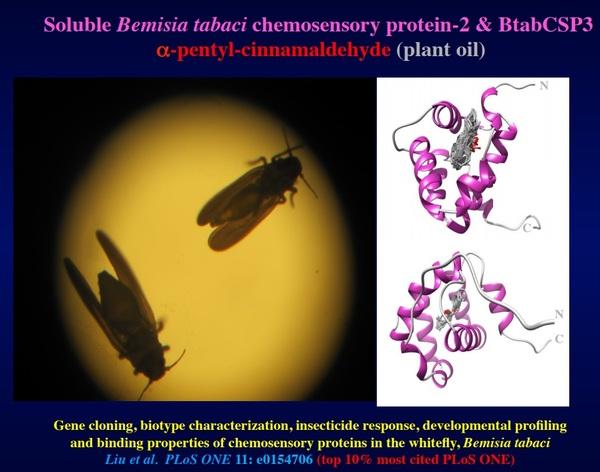
Figure 5. Knock out of BtabCSP2 & CSP3 for inhibition of whitefly detoxifies plant defense. Three-dimensional structure of Bemisia tabaci chemosensory protein-2 &3 bound to α-pentyl-cinnamaldehyde (in grey). Use of derivative of α-pentyl cinnamic aldehyde as ORSA for CSP-based integrated pest management and whitefly control strategy following Liu et al. -2019 (“Pheromone, natural odor and odorant reception suppressing agent (ORSA) for insect control”, Chapter 12, Olfactory Concepts of Insect Control - Alternative to Insecticides, Springer Nature Switzerland AG, 2019). Using Cinnamaldehyde ORSA and analogous compound of plant origin may block the functional binding sites of CSP2 and CSP3 thereby interfering with detoxification mechanisms of whiteflies to defuse plants defense. The insect becomes vulnerable unable to survive the natural toxins from plant oils

Related topics:
Authors:
Recommend
Comment
Share

Would you like to discuss another topic? Create a new post to engage with experts in the community.






.jpg&w=3840&q=75)