Antimicrobial resistance in animal health
Tetracycline Resistance Profile in Darwin Finches in the Galapagos Islands
Author details:
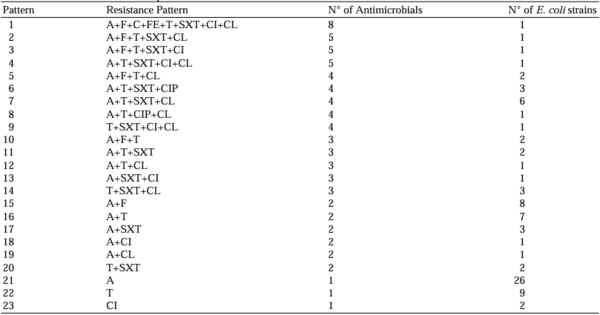
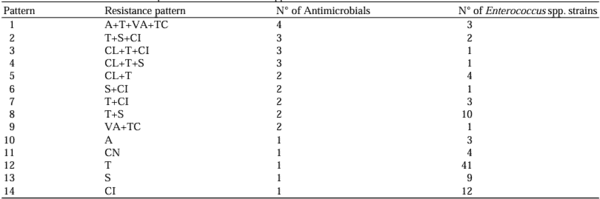
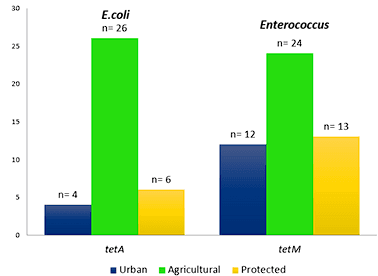
Alonso, C. A., de Toro, M., de la Cruz, F., & Torres, C. (2021). Genomic insights into drug resistance and virulence platforms, CRISPR-Cas systems and phylogeny of commensal E. Coli from wildlife.
Microorganisms, 9(5), 999. https://doi.org/10.3390/microorganisms9050999
Atterby, C., Ramey, A. M., Hall, G. G., Järhult, J.,
Börjesson, S., & Bonnedahl, J. (2016). The increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments.
Infection
Epidemiology, 6(1), 32334. https://doi.org/10.3402/iee.v6.32334
Ecology &
Blanco, G., Lemus, J. A., Grande, J., Gangoso, L.,
Grande, J. M., Donázar, J. A., ... & Hiraldo, F. (2007). Retracted Geographical variation in cloacal microflora and bacterial antibiotic resistance in a threatened avian scavenger concerning diet and livestock farming practices.
Environmental Microbiology, 9(7), 1738-1749. https://doi.org/10.1111/j.1462-2920.2007.01291.x
Bonnedahl, J., & Järhult, J. D. (2014). Antibiotic resistance in wild birds. Upsala Journal of Medical
Sciences, 119(2), 113-116. https://doi.org/10.3109/03009734.2014.905663
CLSI. (2019).
Performance
Standards for
Antimicrobial Susceptibility Testing - 29th Edition (Clinical and Laboratory Standards Institute
[CLSI] (ed.); 29th ed.). CLSI.
Cochetti, I., Vecchi, M., Mingoia, M., Tili, E., Catania, M.
R., Manzin, A., ... & Montanari, M. P. (2005).
Molecular characterization of pneumococci with efflux mediated erythromycin resistance and identification of a novel mef gene subclass, mef (I). Antimicrobial
Agents and Chemotherapy, 49(12), 4999-5006. https://doi.org/10.1128/AAC.49.12.4999-5006.2005
Foti, M., Mascetti, A., Fisichella, V., Fulco, E., Orlandella,
B. M., & Lo Piccolo, F. (2017). Antibiotic resistance assessment in bacteria isolated in migratory
Passeriformes transiting through the Metaponto territory (Basilicata, Italy). Avian Research, 8(1), 1-11. https://doi.org/10.1186/s40657-017-0085-2
Frazzon, A. P. G., Gama, B. A., Hermes, V., Bierhals, C.
G., Pereira, R. I., Guedes, A. G., ... & Frazzon, J. (2010). Prevalence of antimicrobial resistance and molecular characterization of tetracycline resistance mediated by tet (M) and tet (L) genes in Enterococcus spp. isolated from food in Southern Brazil. World
Journal of Microbiology and Biotechnology, 26(2),
365-370. https://doi.org/10.1007/s11274-009-0160-x
Gambino, D., Vicari, D., Vitale, M., Schirò, G., Mira,
F., Giglia, M. L., ... & Gargano, V. (2021). Study on Bacteria Isolates and Antimicrobial Resistance in
Wildlife in
Sicily,
Microorganisms, 9(1), 203.
Southern
Italy. https://doi.org/10.3390/microorganisms9010203
Giacopello, C., Foti, M., Mascetti, A., Grosso, F.,
Ricciardi, D., Fisichella, V., & Lo Piccolo, F. (2016).
Antibiotico resistenza in ceppi di Enterobacteriaceae isolati da avifauna europea ricoverata presso un centro di recupero per la fauna selvatica. Vet. Ital, 52,
139-144. https://doi.org/10.12834/VetIt.327.1374.2
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harbor Perspectives in
Medicine, 6(4), a025387. https://doi.org/10.1101/cshperspect.a025387
Guenther, S., Grobbel, M., Lübke-Becker, A., Goedecke,
A., Friedrich, N. D., Wieler, L. H., & Ewers, C. (2010). Antimicrobial resistance profiles of
Escherichia coli from common European wild bird species. Veterinary Microbiology, 144(1-2), 219-225. https://doi.org/10.1016/j.vetmic.2009.12.016
Guo, T., Lou, C., Zhai, W., Tang, X., Hashmi, M. Z.,
Murtaza, R., ... & Xu, J. (2018). Increased occurrence of heavy metals, antibiotics, and resistance genes in surface soil after long-term application of manure.
Science of the Total Environment, 635, 995-1003. https://doi.org/10.1016/j.scitotenv.2018.04.194
Handrova, L., & Kmet, V. (2019). Antibiotic resistance and virulence factors of Escherichia coli from eagles and goshawks. Journal of Environmental Science and Health, Part B, 54(7), 605-614. https://doi.org/10.1080/03601234.2019.1608103
Hau, M., & Wikelski, M. (2001). Darwin's Finches. Life
Sciences, January, 1–8. https://doi.org/10.1038/npg.els.0001791
Hudzicki, J. (2009). Kirby-Bauer disk diffusion susceptibility test protocol. American Society for
Microbiology, 15, 55-63.
Klibi, N., Ben Amor, I., Rahmouni, M., Dziri, R., Douja, G.,
Ben Said, L., ... & Torres, C. (2015). Diversity of species and antibiotic resistance among fecal enterococci from wild birds in Tunisia. Detection of vanA-containing
Enterococcus faecium isolates. European Journal of
Wildlife Research, 61(2), 319-323. https://doi.org/10.1007/s10344-014-0884-2
Knutie, S. A., Chaves, J. A., & Gotanda, K. M. (2019).
Human activity can influence the gut microbiota of
Darwin's finches in the Galapagos Islands.
Molecular Ecology, 28(9), 2441-2450. https://doi.org/10.1111/mec.15088
Lee, J., Shin, S. G., Jang, H. M., Kim, Y. B., Lee, J., & Kim,
Y. M. (2017). Characterization of antibiotic resistance genes in representative organic solid wastes: Food waste-recycling wastewater, manure, and sewage sludge. Science of the Total Environment, 579, 1692
1698. https://doi.org/10.1016/j.scitotenv.2016.11.187
Lima, T., Domingues, S., & Da Silva, G. J. (2020).
Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events.
Veterinary Sciences, 7(3), 110. https://doi.org/10.3390/vetsci7030110
Liu, C., Chen, Y., Li, X., Zhang, Y., Ye, J., Huang, H., &
Zhu, C. (2020). Temporal effects of repeated application of biogas slurry on soil antibiotic resistance genes and their potential bacterial hosts.
Environmental Pollution, 258, 113652. https://doi.org/10.1016/j.envpol.2019.113652
Lopardo, H. Á. (2016). Introducción a la microbiología clínica. Libros de Cátedra. http://sedici.unlp.edu.ar/handle/10915/52389
Marosevic, D., Kaevska, M., & Jaglic, Z. (2017).
Resistance to the tetracyclines and macrolide lincosamide-streptogramin group of antibiotics and its genetic linkage–a review. Annals of Agricultural and Environmental Medicine, 24(2), 338. https://doi.org/10.26444/aaem/74718
Marston, H. D., Dixon, D. M., Knisely, J. M., Palmore, T.
N., & Fauci, A. S. (2016). Antimicrobial resistance. Jama, 316(11), 1193-1204. https://doi.org/10.1001/jama.2016.11764
Michalova, E., & Schlegelova, J. (2004). Tetracyclines in veterinary medicine and bacterial resistance to them. Veterinarni Medicina, 49(3), 79. https://doi.org/10.17221/5681-VETMED
Michel, A. J., Ward, L. M., Goffredi, S. K., Dawson,
K. S., Baldassarre, D. T., Brenner, A., ... & Chaves,
J. A. (2018). The gut of the finch: uniqueness of the gut microbiome of the Galápagos vampire finch. Microbiome, 6(1), 1-14. https://doi.org/10.1186/s40168-018-0555-8
Millar, B. C., Jiru, X. U., Moore, J. E., & Earle, J. A. (2000).
A simple and sensitive method to extract bacterial, yeast and fungal DNA from blood culture material. Journal of
Microbiological Methods, 42(2), 139-147. https://doi.org/10.1016/S0167-7012(00)00174-3
Nieto-Claudin, A., Deem, S. L., Rodríguez, C., Cano, S.,
Moity, N., Cabrera, F., & Esperón, F. (2021).
Antimicrobial resistance in Galapagos tortoises as an indicator of the growing human footprint.
Environmental Pollution, 284, 117453. https://doi.org/10.1016/j.envpol.2021.117453
Nowaczek, A., Dec, M., Stępień-Pyśniak, D., Urban
Chmiel, R., Marek, A., & Różański, P. (2021).
Antibiotic Resistance and Virulence Profiles of
Escherichia coli Strains Isolated from Wild Birds in
Poland. Pathogens, 10(8), 1059. https://doi.org/10.3390/pathogens10081059
Pantozzi, F. L. (2018). Evaluación fenotípica y genotípica de la resistencia a tetraciclina en cepas de
Escherichia coli de origen animal (Doctoral dissertation, Universidad Nacional de La Plata). http://sedici.unlp.edu.ar/handle/10915/70535
Radhouani, H., Poeta, P., Goncalves, A., Pacheco, R., Sargo,
R., & Igrejas, G. (2012). Wild birds as biological indicators of environmental pollution: Antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). Journal of
Medical
Microbiology,
61(6), https://doi.org/10.1099/jmm.0.038364-0
837-843.
Radimersky, T., Frolkova, P., Janoszowská, D., Dolejská,
M., Svec, P., Roubalová, E., ... & Literák, I. (2010).
Antibiotic resistance in fecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. Journal of
Applied Microbiology, 109(5), 1687-1695. https://doi.org/10.1111/j.1365-2672.2010.04797.x
Ramey, A. M., & Ahlstrom, C. A. (2020). Antibiotic resistant bacteria in wildlife: Perspectives on trends, acquisition and dissemination, data gaps and future directions. Journal of Wildlife Diseases, 56(1), 1-15. https://doi.org/10.7589/2019-04-099
Rossolini, G. M., Arena, F., & Giani, T. (2017).
Mechanisms of Antibacterial Resistance. In
Infectious Diseases (Fourth Edi). Elsevier Ltd. https://doi.org/10.1016/b978-0-7020-6285-8.00138-6
Santos, T., Silva, N., Igrejas, G., Rodrigues, P., Micael, J.,
Rodrigues, T., ... & Poeta, P. (2013). Dissemination of antibiotic-resistant Enterococcus spp. and
Escherichia coli from wild birds of Azores
Archipelago. Anaerobe, 24, 25-31. https://doi.org/10.1016/j.anaerobe.2013.09.004
Sarmah, A. K., Meyer, M. T., & Boxall, A. B. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate, and effects of veterinary antibiotics (VAs) in the environment. Chemosphere,
65(5), 725-759. https://doi.org/10.1016/j.chemosphere.2006.03.026
Schell, C. M. B. (2019). Caracterización fenotípica y genotípica de cepas de Enterococcus spp. aisladas de líquidos obtenidos por punción provenientes de infecciones invasivas humanas (Doctoral dissertation, Universidad Nacional de La Plata). http://sedici.unlp.edu.ar/handle/10915/73675
Sigirci, B. D., Celik, B., Kahraman, B. B., Bagcigil, A. F., &
Ak, S. (2019). Tetracycline resistance of enterobacteriaceae isolated from feces of synanthropic birds. Journal of Exotic Pet Medicine, 28, 13-18. https://doi.org/10.1053/j.jepm.2017.12.003
Silva, V., Igrejas, G., Carvalho, I., Peixoto, F., Cardoso,
L., Pereira, J. E., ... & Poeta, P. (2018). Genetic
Characterization of van A-Enterococcus faecium
Isolates from Wild Red-Legged Partridges in
Portugal. Microbial Drug Resistance, 24(1), 89-94. https://doi.org/10.1089/mdr.2017.0040
Skarżyńska, M., Zajac, M., Bomba, A., Bocian, Ł., Kozdruń,
W., Polak, M., ... & Wasyl, D. (2021). Antimicrobial
Resistance Glides in the Sky-Free-Living Birds as a
Reservoir of Resistant Escherichia coli With Zoonotic
Potential. Frontiers in Microbiology, 12, 656223. https://doi.org/10.3389/fmicb.2021.656223
Stępień-Pyśniak, D., Hauschild, T., Dec, M., Marek, A.,
& Urban-Chmiel, R. (2019). Clonal structure and antibiotic resistance of Enterococcus spp. from wild birds in Poland. Microbial Drug Resistance, 25(8),
1227-1237. https://doi.org/10.1089/mdr.2018.0461
Thaker, M., Spanogiannopoulos, P., & Wright, G. D. (2010). The tetracycline resistome. Cellular and
Molecular Life Sciences, 67(3), 419-431. https://doi.org/10.1007/s00018-009-0172-6
UNESCO. (2022). Galápagos Islands. https://whc.unesco.org/en/list/1/
Wall, B. A., Mateus, A. L. P., Marshall, L., Pfeiffer, D.
U., Lubroth, J., Ormel, H. J., ... & Patriarchi, A. (2016). Drivers, dynamics, and epidemiology of antimicrobial resistance in animal production. Food and Agriculture Organization of the United Nations.
Wheeler, E., Hong, P. Y., Bedon, L. C., & Mackie, R. I. (2012). Carriage of antibiotic-resistant enteric bacteria varies among sites in Galapagos reptiles. Journal of Wildlife Diseases, 48(1), 56-67. https://doi.org/10.7589/0090-3558-48.1.56
White, A., & Hughes, J. M. (2019). The critical importance of a one health approach to antimicrobial resistance. Eco Health, 16(3), 404-409. https://doi.org/10.1007/s10393-019-01415-5
WHO. (2018). WHO|WHO list of Critically Important
Antimicrobials (CIA).
WHO/AGISAR. (2017). Integrated Surveillance of
Antimicrobial Resistance in Foodborne Bacteria
Application of a One Health Approach.
Xiong, W., Wang, M., Dai, J., Sun, Y., & Zeng, Z. (2018).
The application of manure containing tetracyclines slowed down the dissipation of tet-resistance genes and caused changes in the composition of soil bacteria.
Ecotoxicology and Environmental Safety, 147, 455-460. https://doi.org/10.1016/j.ecoenv.2017.08.061
Yahia, H. B., Chairat, S., Hamdi, N., Gharsa, H., Sallem, R.
B., Ceballos, S., ... & Slama, K. B. (2018).
Antimicrobial resistance and genetic lineages of fecal enterococci of wild birds: Emergence of vanA and vanB2 harboring Enterococcus faecalis. International
Journal of Antimicrobial Agents, 52(6), 936-941. https://doi.org/10.1016/j.ijantimicag.2018.05.005










.jpg&w=3840&q=75)