INTRODUCTION
Salmonella is an important pathogen transmitted through food, water or direct contact with animals. Recently, it was estimated that this pathogen causes more than 20 million disease episodes and 144,000 worldwide deaths annually.1 In order to prevent infections, veterinary and human health laboratories have stablished national Salmonella surveillance programs oriented to monitor prevalence, distribution and antibiotic resistance profiles of this pathogen.2,3 The majority of these programs rely on PCR-based assays for the rapid and accurate detection of Salmonella spp.3-5
Numerous PCR assays have been developed for molecular detection of Salmonella spp., many of them revealing different levels of specificity and accuracy.3,6,7 Among these molecular tools, the invA-based PCR assay has been accepted as the conventional method for detection of Salmonella spp. in animal and human clinical samples.3,8-10 This PCR protocol was originally proposed by Rahn et al. in 199211 and amplifies a 284-bp DNA fragment of the invA gene, a Salmonella-specific locus. This PCR assay has been extensively validated for its use as an international standard tool for accurate detection of Salmonella spp.3,8-10 Although the invA-based PCR assay is now considered one of the standard methods for detection of Salmonella spp., numerous reports have described the occurrence of false-positive results in PCR reactions using DNA obtained from non-Salmonella isolates.3,4,10-12 To this end, our research group has recovered multiple Citrobacter spp. isolates from poultry meat samples in which the standard invAbased PCR assay3 generates false-positive results. Thus, the main goal of the present study was to standardize a rapid, feasible, and accurate PCR protocol for the detection of Salmonella isolates.
MATERIAL AND METHODS
Bacterial Isolates and Culture Conditions
Salmonella enterica type-strains (Typhimurium ATCC-14028 and Typhi ATCC-6539) and non-type-strains (laboratory collection), as well as Citrobacter spp. isolates (laboratory collection) were retrieved from our laboratory frozen-glycerol stock collection. The laboratory collection comprises > 200 bacterial isolates obtained during a Poultry Meat Salmonella Surveillance Program. Bacteria strains were grown overnight in tryptic soy broth (BD Difco, Mexico) at 35°C. After incubation, one milliliter of the culture was used for genomic DNA extraction using a commercial kit (Quick-DNA Miniprep Plus Kit, Irvine, CA) following manufacturer instructions. DNA quantity and quality were determined using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE).
PCR Assay Targeting the invA Gene
Genomic DNA extracted from S. enterica and Citrobacter spp. isolates were sub jected to invA-PCR amplification using the protocol described for primers invA-139 and invA-141.3 Also, an alternative PCR assay with primers invA-1 and invA-213 was evaluated (Table 1). When published PCR protocols generated non-specific amplicons using DNA from Citrobacter spp. as a template, gradient PCR runs (temperature range: 41 to 64 °C) were performed to establish optimum annealing temperature. All PCR reactions were carried out with Maxima Hot Start Taq DNA Polymerase (Thermo Fisher Scientific, Waltham, MA) as described by the manufacturer.
The optimized PCR protocol used with primers invA-139 + invA-141 consisted of an initial denaturation at 94°C 60 s, 35 cycles of: 94°C 30 s, 64°C 30 s, 72°C 30 s, and a final extension step at 72°C for 4 min. The optimized PCR protocol used with primers invA-1 + invA-2 consisted of an initial denaturation at 94°C 3 min, 35 cycles of: 94°C 30 s, 57.4°C 30 s, 72°C 30 s, and a final extension step at 72°C for 5 min. Specificity of the PCR method was confirmed by visualizing single bands corresponding to DNA fragments of the expected size via ethidium bromide/agarose gel electrophoresis.
Complementary Salmonella-specific PCR Assays
To improve the discriminatory power of the invA-based PCR protocol, alternative Salmonella-specific PCR assays were evaluated. Three additional primers sets: (16SF1 + 16SIII, MINf + MINr, and ttr-6 + ttr-4) were evaluated using protocols published elsewhere14-16 (Table 1). Similarly, when published PCR protocols generated nonspecific amplicons, gradient PCR runs (temperature range: 41 to 64 °C) were performed to establish optimum annealing temperature. The optimized PCR protocol used with primers MINf + MINr consisted of an initial denaturation at 94°C 3 min, 32 cycles of: 94°C 20 s, 53°C 30 s, 72°C 30 s, and a final extension step at 72°C for 2 min. Specificity of the PCR method was confirmed by visualizing single bands corresponding to DNA fragments of the expected size via ethidium bromide/agarose gel electrophoresis.
16S rRNA Gene Sequencing and Phylogenetic Analysis
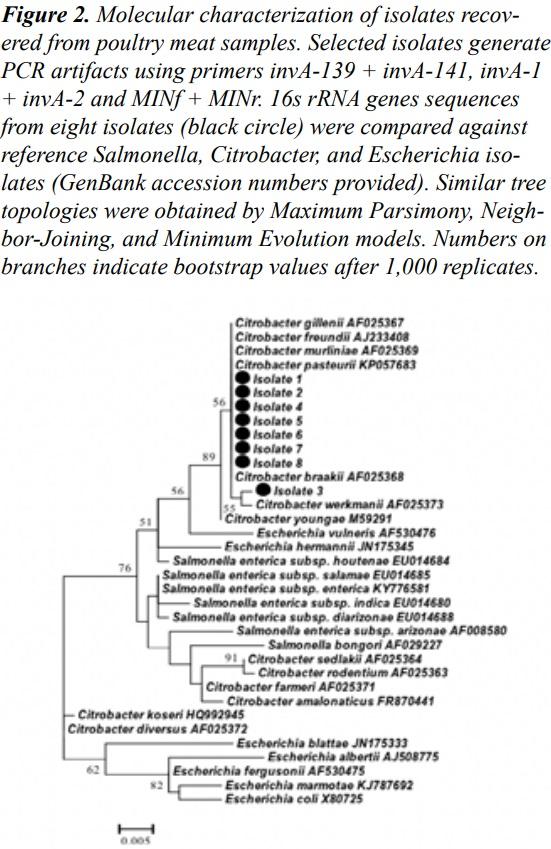
Bacterial isolates identified as non-Salmonella by the invA and 16S rRNA PCR assays were subject to molecular identification by means of PCR amplification of near full-length 16S rRNA gene as described elsewhere.17 PCR products were purified and subjected to Sanger sequencing using an ABI 3730XL capillary sequencer. Inspection, alignment, and trimming of sequences were performed with MEGA6 software.18 Initial identification was performed using the Ribosomal Database Project (RDP) Classifier and Sequence Match tools.19 Then, a phylogenetic analysis was performed using 16S rRNA genes from Salmonella and Citrobacter species described in the Approved Lists of Bacterial Names published.20 Phylogenetic inference by means of Maximum Parsimony (MP), Neighbor-Joining (NJ), and Minimum Evolution (ME) tree models was estimated with the MEGA6 software.18 Statistical significance of branch order was estimated by bootstrap analysis with 1,000 replicates.
RESULTS
PCR Assay Targeting the invA Gene
A series of PCR runs were carried out to validate the specificity of the invA-based PCR assays using protocols published elsewhere.3,13 Under our laboratory conditions, published protocols using primers invA-139 + invA-141 and invA-1 + invA-2 generated non-specific signals in reactions containing Citrobacter DNA (data not shown). Therefore, new PCR conditions were established by using temperature gradient assays. After numerous efforts, it was not possible to eliminate non-specific signals with primer pair invA-139 and invA-141. In contrast, specify of primers invA-1 and invA-2 was enhanced with an optimized PCR protocol (Figure 1A).
Complementary Salmonella-specific PCR Assays
Due to the occurrence of non-specific amplifications (false-positive results) with the invA-based PCR assay, it was decided to establish a double-PCR protocol for accurate detection S. enterica. After multiple attempts, primers pairs 16SF1 + 16SIII and ttr-6 + ttr-4 generated non-specific signals in reactions using Citrobacter DNA. Importantly, it was possible to eliminate these false-positive reactions with an optimized PCR protocol using primers MINf + MINr (Figure 1A).
Figure 1. PCR assays for detection of Salmonella spp. Representative PCR reactions using isolates recovered from poultry meat samples. A) Salmonella enterica and Citrobacter spp. isolates were used for PCR primer validation. B) Double PCR approach for detection of Salmonella spp. With this approach, Salmonella isolates produce single, artifact-free, and size-expected amplicons. Citrobacter spp. isolates were subjected to molecular identification (Figure 2).
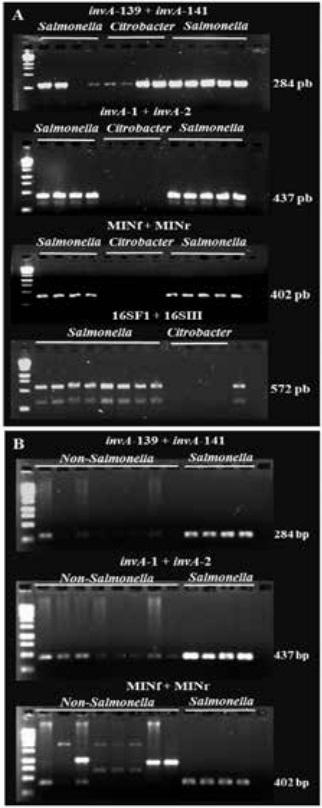
To evaluate the effectiveness of this double-PCR protocol for accurate detection S. enterica, a collection of > 200 poultry meat isolates was subjected to PCR amplification using primers invA-139 + invA-141, invA-1 + invA-2, and MINf + MINr. In all PCR assays, non-specific amplifications (weak amplicons and artifacts) were observed in numerous reactions. However, PCR reactions containing DNA extracted from confirmed S. enterica produced DNA fragments of the expected size and free of PCR artifacts (Figure 1B). To corroborate this observation, representative isolates generating non-specific amplifications (weak amplicons) and artifacts were characterized by sequencing of the 16S rRNA gene.
Molecular Identification by Means of 16S rRNA Gene Phylogenetic Analysis
Initial characterization performed by RDP Classifier and Sequence Match tools19 identified the selected isolates as members of the genus Citrobacter (data not shown). This result was confirmed by a phylogenetic analysis using Maximum Parsimony (MP), Neighbor-Joining (NJ), and Minimum Evolution (ME) tree models. Together, these phylogenetic analyses confirmed that isolates generating non-specific amplifications and artifacts belonged to the genus Citrobacter (Figure 2). The 16S rRNA gene sequences obtained in the present study were deposited at the GenBank under accession numbers: MG597049-MG597056.
DISCUSSION
Worldwide, numerous veterinary and human health laboratories rely on the invA-based PCR assay for detection of Salmonella spp. in clinical samples.3,8-10 Despite the fact that this PCR assay is considered one of the standard methods for detection of Salmonella spp., numerous reports have documented false-positive results caused by the appearance of non-specific amplifications and artifacts in the PCR reactions.3,4,10-12
As reported elsewhere, published PCR protocols with primers invA-139 + invA-141 and invA-1 + invA-2 generate non-specific signals in reactions containing DNA from non-Salmonella strains.3,4,10-12 To improve the discriminatory power of invA-based PCR protocols, we evaluated the performance of three validated Salmonella-specific primers targeting 16S rRNA and functional genes.14-16 Using Citrobacter isolates selected by their known capability of generating false-positive results, it was revealed that primers MINf + MINr were more reliable for detecting S. enterica isolates. This superior specificity was also demonstrated using a set of 78 S. enterica strains representing 31 different serovars, and 23 non-Salmonella strains.16 Thus, it is recommended to perform detection of Salmonella species by using the combination of PCR protocols targeting the invA gene and the selected locus of the 16S rRNA gene.
The performance of this double PCR approach was evaluated using a large set of bacterial isolates recovered from poultry meat. The analysis revealed that S enterica isolates generate single, artifact-free, and size-expected amplicons. In contrast, non-Salmonella isolates produce weak amplicons and a series of non-specific PCR products. Interestingly, all selected non-Salmonella strains belonged to the genus Citrobacter. We have continued the design and evaluation of alternative PCR protocols for the accurate detection of S. enterica. However, so far, none of the evaluated primer sets have been able to eliminate the nonspecific amplification generated by these challenging Citrobacter isolates.
CONCLUSION
For the rapid, feasible and accurate detection of Salmonella isolates, it is recommended to use of a double PCR approach, using primes targeting the invA (invA-1 + invA-2) and 16S rRNA (MINf + MINr) genes. Under these assays, S enterica isolates produce single, artifact-free, and size-expected amplicons, which are easily distinguishable from nonSalmonella isolates due to the generation of weak amplicons and PCR artifacts.
This article was originally published in Intern J Appl Res Vet Med • Vol. 16, No. 2, 2018.













.jpg&w=3840&q=75)