Identification of parental line specific effects of MLF2 on resistance to coccidiosis in chickens
Background: MLF2 was the candidate gene associated with coccidiosis resistance in chickens. Although single marker analysis supported the association between MLF2 and coccidiosis resistance, causative mutation relevant to coccidiosis was not identified yet. Thus, this study suggested segregation analysis of MLF2 haplotype and the association test of the other candidate genes using improved data transformation.
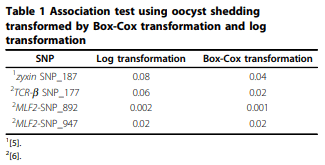
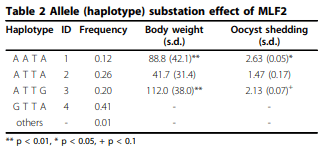
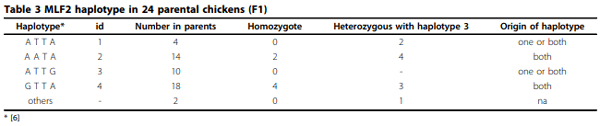
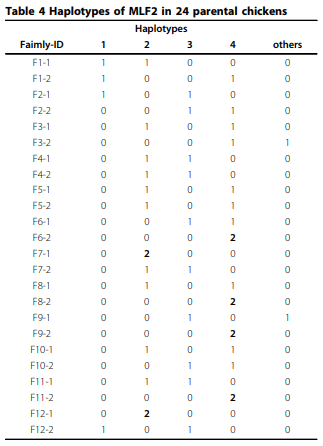
Results: A haplotype probably originated from one parental line was found out of 4 major haplotypes of MLF2. Frequency of this haplotype was 0.2 in parental chickens and its offspring in 12 families. Allele substitution effect of the MLF2 haplotype originated from a specific line was associated with increased body weight and fecal egg count explaining coccidiosis resistance. Nevertheless, Box-Cox transformation was able to improve normality; association test did not produce obvious different results compared with analysis with log transformed phenotype.
Conclusion: Allele substitution effect analysis and classification of MLF2 haplotype identified the segregation of haplotype associated with coccidiosis resistance. The haplotype originated from a specific parental line was associated with improving disease resistance. Estimating effect of MLF2 haplotype on coccidiosis resistance will provide useful information for selecting animals or lines for future study.
1. Lillehoj HS, Kim CH, Keeler CL, Zhang S: Immunogenomic approaches to study host immunity to enteric pathogens. Poultry Science 2007, 86(7):1491-1500.
2. Johnson LW, Edgar SA: Response to prolonged selection for resistance and susceptibility to acute cecal occidiosis in the Auburn strain Single Comb White Leghorn. Poult Sci 1982, 61(12):2344-2355.
3. Zhu JJ, Lillehoj HS, Allen PC, Van Tassell CP, Sonstegard TS, Cheng HH, Pollock D, Sadjadi M, Min W, Emara MG: Mapping quantitative trait loci associated with resistance to coccidiosis and growth. Poultry Science 2003, 82(1):9-16.
4. Kim ES, Hong YH, Min W, Lillehoj HS: Fine-mapping of coccidia-resistant quantitative trait loci in chickens. Poultry Science 2006, 85(11):2028-2030.
5. Hong YH, Kim ES, Lillehoj H, Song KD, Lillehoj E: Association of resistance to avian coccidiosis with single nucleotide polymorphisms in the Zyxin gene. Poultry Science 2009, 88(3):511-518.
6. Kim ES, Hong YH, Lillehoj HS: Genetic effects analysis of myeloid leukemia factor 2 and T cell receptor-beta on resistance to coccidiosis in chickens. Poultry Science 2010, 89(1):20-27.
7. Allen PC: Production of free radical species during Eimeria maxima infections in chickens. Poultry Science 1997, 76(6):814-821.
8. Yun CH, Lillehoj HS, Zhu J, Min W: Kinetic differences in intestinal and systemic interferon-γ and antigen-specific antibodies in chickens experimentally infected with Eimeria maxima. Avian Dis 2000, 44(2):305-312.
9. Box GEP, Cox DR: An analysis of transformations (with discussion). Journal of the Royal Statistical Society B 1964, 26:211-252.
10. Venables WN, Ripley BD: Modern applied statistics with S-plus. Springer; 1999.









.jpg&w=3840&q=75)