Genetic Variation among Avian Pathogenic E. coli Strains Isolated from Broiler Chickens
Background: Avian pathogenic E. coli cause serious disease in chickens, which mainly chracterised by airsaculitis, perihepatitis and pericarditis resulting in large economic losses in poultry industry worldwide.
Methodology: In order to investigate the genetic variation among different E. coli strains isolated from chickens, one hundred diseased broiler chickens at Sharkia province, Egypt were examined. Liver and heart blood samples were collected from each bird and subjected to bacteriological examination, where the prevalence of E. coli was 60% from the total collected samples. Escherichia coli strains were serogrouped. The PCR was used for detection of Shiga-like toxins genes (stx1 and stx2), attaching and effacing (eaeA) gene and enterohaemolysin gene (hly) in the typable isolated E. coli strains.
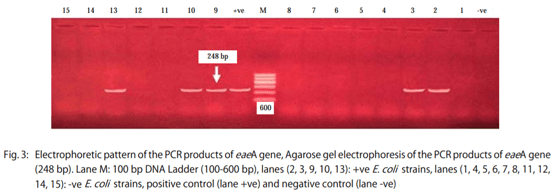
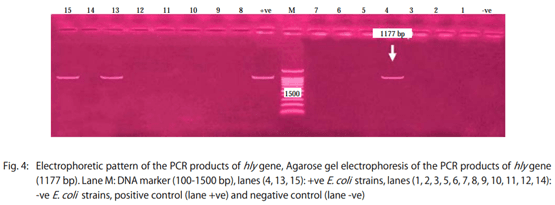
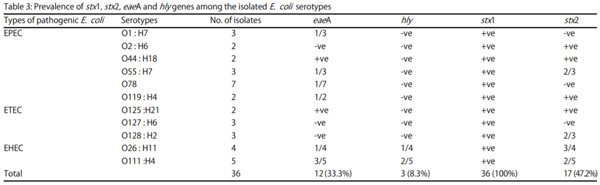
Results: The results showed that the isolated strains belonged to 11 serogroups including O1, O2, O26, O44, O55, O78, O111, O119, O125, O127 and O128. Untypable strains were also recovered. The detected virulence genes were stx1 in all E. coli strains (100%), stx2 in 17 strains (47.2%), eaeA in 12 strains (33.3%) and hly only in three strains (8.3%).
Conclusion: In conclusion, the combination of genotypic and phenotypic analysis of E. coli is more valuable as an epidemiological tool for identification of isolates. This study established the presence of stx1 and stx2 containing E. coli in chickens.
Key words: Avian pathogenic E. coli, broilers, virulence genes, PCR
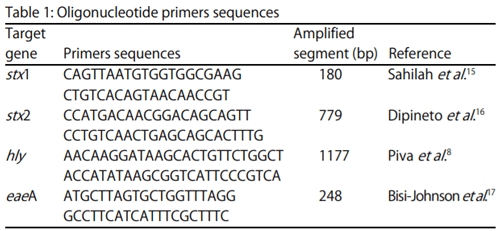
- The stx1 gene according to Sahilah et al. 15: Initial denaturation at 94°C for 7 min, 35 cycles (denaturation at 94°C for 30 sec, annealing at 51°C for 30 sec, extension at 72°C for 30 sec), final extension 72°C for 7 min
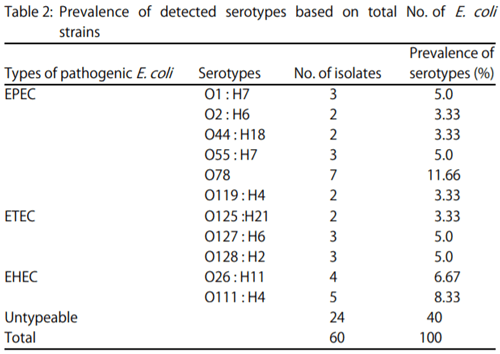
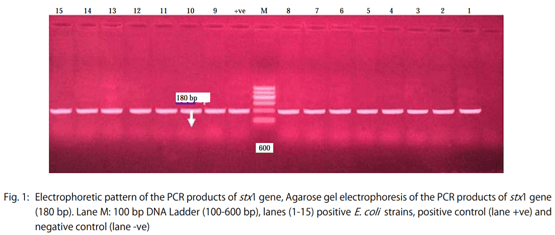
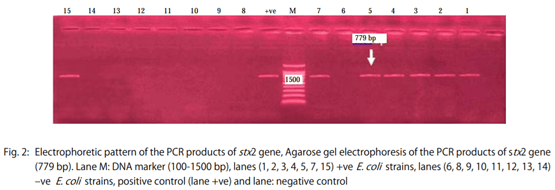
- The stx2 gene according to Dipineto et al. 16: Initial denaturation at 94°C for 10 min, 35 cycles (denaturation at 94°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 1 min), final extension 72°C for 10 min.
- The hly gene according to Piva et al. 8 : Initial denaturation at 94°C for 15 min, 35 cycles (denaturation at 94°C for 1 min, annealing at 60°C for 1 min, extension at 72°C for 15 min), final extension 72°C for 12 min.
- The eaeA gene according to Bisi-Johnson et al. 17: Initial denaturation at 94°C for 7 min, 40 cycles (denaturation at 94°C for 30 sec, annealing at 51°C for 30 sec, extension at 72°C for 30 sec), final extension 72°C for 7 min.
1. Samah Eid, A.S. and A.M. Erfan, 2013. Characterization of E. coli associated with high mortality in poultry flocks. Assiut Vet. Med. J., 59: 51-61.
2. Tivendale, K.A., C.M. Logue, S. Kariyawasam, D. Jordan and A. Hussein et al., 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immunity, 78: 3412-3419.
3. Xia, X., J. Meng, P.F. McDermott, S. Ayers and K. Blickenstaff et al., 2010. Presence and characterization of Shiga toxin-producing Escherichia coli and other potentially diarrheagenic E. coli strains in retail meats. Applied Environ. Microbiol., 76: 1709-1717.
4. Abong'o, B.O. and M.N. Momba, 2009. Prevalence and characterization of Escherichia coli O157:H7 isolates from meat and meat products sold in Amathole District, Eastern cape province of South Africa. Food Microbiol., 26: 173-176.
5. Karch, H., P.I. Tarr and M. Bielaszewska, 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol., 295: 405-418.
6. Elfeil, W., E. Soliman and M. Sobeih, 2011. Epidemiological Studies on Environmental Pollution in Poultry Farms. GRIN Publishing, Munich, Germany.
7. Caprioli, A., S. Morabito, H. Brugere and E. Oswald, 2005. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet. Res., 36: 289-311.
8. Piva, I.C., A.L. Pereira, L.R. Ferraz, R.S.N. Silva and A.C. Vieira et al., 2003. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasilia, Brazil. J. Clin. Microbiol., 41: 1827-1832.
9. Quinn, P.J., B.K. Markey, M.E. Carter, W.J. Donnelly and F.C. Leonard, 2002. Veterinary Microbiology and Microbial Diseases. 1st Edn., Wiley-Blackwell Science, USA., ISBN-13: 978-0632055258, Pages: 544.
10. Edwards, P.R. and W.H. Ewing, 1972. Identification of Enterobacteriaceae. 3rd Edn., Burgess Publishing Co., Minnecepolis, Minnesota, pp: 103-104.
11. Davies, R.H. and C. Wray, 1997. Immunomagnetic separation for enhanced flagellar antigen phase inversion in salmonella. Lett. Applied Microbiol., 24: 217-220.
12. Berkhoff, H.A. and A.C. Vinal, 1986. Congo red medium to distinguish between invasive and non-invasive Escherichia coli pathogenic for poultry. Avian Dis., 30: 117-121.
13. Livezey, B.C. and R.L. Zusi, 2007. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zool. J. Linnean Soc., 149: 1-95.
14. Sambrook, J. and D.W. Russell, 2001. Molecular Cloning: A Laboratory Manual. 3rd Edn., Cold Spring Harbor Laboratory Press, New York, USA., ISBN-13: 9780879695774, Pages: 2344.
15. Sahilah, A.M., L.Y.Y. Audrey, S.L. Ong, W.N. Wan Sakeenah and S. Safiyyah et al., 2010. DNA profiling among egg and beef meat isolates of Escherichia coli by Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR) and Random Amplified Polymorphic DNA-PCR (RAPD-PCR). Int. Food Res. J., 17: 853-866.
16. Dipineto, L., A. Santaniello, M. Fontanella, K. Lagos, A. Fioretti and L.F. Menna, 2006. Presence of Shiga toxin-producing Escherichia coli O157:H7 in living layer hens. Lett. Applied Microbiol., 43: 293-295.
17. Bisi-Johnson, M.A., C.L. Obi, S.D. Vasaikar, K.A. Baba and T. Hattori, 2011. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog, Vol. 3.
18. Abouelmaatti, R.R., A.M. Algammal, X. Li, J. Ma, E.A. Abdelnaby and W.M.K. Elfeil, 2013. Experimental immunology cloning and analysis of Nile tilapia Toll-like receptors type-3 mRNA. Cent. Eur. J. Immunol., 38: 277-282.
19. Peer, F.U., M.M. Ansari, I.A. Gani and M.M. Willayat, 2013. Serotyping and antibiotic sensitivity patterns of Escherchia coli isolates obtained from broiler chicks in Kashmir Valley, India. Adv. Anim. Vet. Sci., 1: 75-76.
20. Ammar, A.M.A.E.H., S.E.A. Eid and A.M. Eloksh, 2014. Studies on virulence genes of E. coli from different sources and their relation to antibiotic resistance pattern. Zagazig Vet. J., 42: 183-196.
21. Literak, I., T. Reitschmied, D. Bujnakova, M. Dolejska and A. Cizeket al., 2013. Broilers as a source of quinolone-resistant and extraintestinal pathogenic Escherichia coli in the Czech Republic. Microb. Drug Resistance, 19: 57-63.
22. Radwan, I.A.E.H., H.S.H. Salam, S.A.E.A. Abd-Alwanis and M.A.Y. Al-Sayed, 2014. Frequency of some virulence associated genes among multidrug-resistant Escherichia coli isolated from septicemic broiler chicken. Int. J. Adv. Res., 2: 867-874.
23. Matthijs, M.G.R., 2008. The Pathogenesis of Colibacillosis in Broilers Infected with Virulent or Vaccine Strains of Infectious Bronchitis Virus. Utrecht University, Netherlands, Pages: 169.
24. Ashraf, A.A.E.T., A.M. Ammar, A.R. Ali, F.I.E. Hofy and M.E.E.S Ahmed, 2013. Detection of common (inv A) gene in salmonellae isolated from poultry using polymerase chain rection technique. Benha Vet. Med. J., 25: 70-77.
25. Hussein, A.H.M., I.A.I. Ghanem, A.A.M. Eid, M.A. Ali and J.S. Sherwood et al., 2013. Molecular and phenotypic characterization of Escherichia coli isolated from broiler chicken flocks in Egypt. Avian Dis., 57: 602-611.
26. Ewers, C., E.M. Antao, I. Diehl, H.C. Philipp and L.H. Wieler, 2009. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Applied Environ. Microbiol., 75: 184-192.
27. Oh, J.Y., M.S. Kang, H. Yoon, H.W. Choi and B.K. An et al., 2012. The embryo lethality of Escherichia coli isolates and its relationship to the presence of virulence-associated genes. Poult. Sci., 91: 370-375.
28. Ozaki, H. and T. Murase, 2009. Multiple routes of entry for Escherichia coli causing colibacillosis in commercial layer chickens. J. Vet. Med. Sci., 71: 1685-1689.
29. Sharda, R., S.W. Ruban and M. Thiyageeswaran, 2010. Isolation, characterization and antibiotic resistance pattern of Escherichia coli isolated from poultry. Am.-Eurasian J. Scient. Res., 5: 18-22.
30. Yoder, Jr. H.W., 1989. Congo red binding by Escherichia coli isolates from chickens. Avian Dis., 33: 502-505.
31. Erganis, O., O. Kaya, M. Corlu and E. Istanbulluoglu, 1989. Hemagglutination, hydrophobicity, enterotoxigenicity and drug-resistance characteristics of avian Escherichia coli. Avian Dis., 33: 631-635.
32. Sharada, R.K., R.G. Raghavan, R.N.S. Gowda and H. Upendra, 1999. Haemagglutination and congo red binding of avian Escherichia coli. Indian J. Comp. Microbiol. Immunol. Infect. Dis., 20: 151-152.
33. Rodriguez, M.F., G.D. Wiens, M.K. Purcell and Y. Palti, 2005. Characterization of Toll-like receptor 3 gene in rainbow trout (Oncorhynchus mykiss). Immunogenetics, 57: 510-519.
34. Momtaz, H. and A. Jamshidi, 2013. Shiga toxin-producing Escherichia coli isolated from chicken meat in Iran: Serogroups, virulence factors and antimicrobial resistance properties. Poult. Sci., 92: 1305-1313.
35. El-Jakee, J.K., R.M. Mahmoud, A.A. Samy, M.A. El-Shabrawy, M.M. Effat and W.A. Gad El-Said, 2012. Molecular characterization of E. coli isolated from chicken, cattle and buffaloes. Int. J. Microbiol. Res., 3: 64-74.
36. Kobayashi, H., T. Pohjanvirta and S. Pelkonen, 2002. Prevalence and characteristics of intimin- and Shiga toxin-producing Escherichia coli from gulls, pigeons and broilers in Finland. J. Vet. Med. Sci., 64: 1071-1073.
37. Schroeder, C.M., D.G. White, B. Ge, Y. Zhang and P.F. McDermott et al., 2003. Isolation of antimicrobial-resistant Escherichia coli from retail meats purchased in Greater Washington, DC, USA. Int. J. Food Microbiol., 85: 197-202.
38. Wani, S.A., I. Samanta, M.A. Bhat and Y. Nishikawa, 2004. Investigation of shiga toxin-producing Escherichia coli in avian species in India. Lett. Applied Microbiol., 39: 389-394.
39. Farah, S.M.S.S., E.M. De Souza, F.O. Pedrosa, K. Irino and L.R. Da Silva et al., 2007. Phenotypic and genotypic traits of Shiga toxin-producing Escherichia coli strains isolated from beef cattle from Parana State, Southern Brazil. Lett. Applied Microbiol., 44: 607-612.
40. Bailey, M.J.A., V. Koronakis, T. Schmoll and C. Hughes, 1992. Escherichia coli HIyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH (sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol. Microbiol., 6: 1003-1012.









.jpg&w=3840&q=75)