Introduction
Exosomes are membrane vesicles, approximately 40–100 nm in diameter, and present in most biological fuids [1–3]. Exosomes are derived from multivesicular bodies (MVBs) to form intraluminal vesicles (ILVs), which are then released into the extracellular environment as exosomes after fusion with the plasma membrane [3]. Lipids and proteins are the main components of exosomes and various nucleic acids, such as mRNAs, microRNAs (miRNAs), and other non-coding RNAs (ncRNAs), and are also found in the exosomal lumen [1– 4]. These exosomal RNAs can be delivered from donor cells to recipient cells, wherein they modulate various biological systems [5–7]. Therefore, exosomes are important in cell-to-cell communication.
miRNAs are small ncRNA molecules and are typically 22 nucleotides in length, which repress translation of the target mRNA by binding to the 3′-untranslated region and/or induce the decay of up to 30% of all expressed transcripts [8]. miRNAs are involved in diverse biological processes, including fat metabolism; cell death, proliferation, differentiation; and the functioning of the immune system [9]. Therefore, exosomal miRNAs from donor cells can regulate the gene expression of recipient cells. Furthermore, the composition of exosomal miRNAs is different between healthy and diseased individuals [10]. Thus, exosomal miRNAs indicate the state of disease and control the immune systems.
Avian influenza viruses (AIV) in Influenza virus A genus, belonging to the family Orthomyxoviridae, cause severe outbreaks in the poultry industry. In particular, the H5N1 subtype is a highly pathogenic avian influenza virus (HPAIV) that originated from Asia [11]. H5N1 in poultry decreases egg production and causes rhinorrhoea, loss of appetite, soft-shelled or misshapen eggs, diarrhoea, and sudden death [12]. Thus, H5N1 outbreak causes significant economic damage to the poultry industry. Furthermore, H5N1 can be transmitted to humans and causes severe acute respiratory infection with a fatality rate of more than 50% [13]. Therefore, it is essential to elucidate the mechanisms of AIV pathogenesis in chickens to control the infection.
Exosomes have various roles in immune response by delivering exosomal contents to other cells such as membrane-bound receptors and miRNAs [14]. Therefore, we suggest that exosomes might have important roles, especially delivering miRNAs, in immune response against infection of H5N1 HPAIV. To date, however, there are no studies describing exosomal small RNA transcriptome analysis in AIV infection. In this study, Ri chickens, local yellow-feathered household chickens of Vietnam [15], were used as experimental animals. Further, by genotyping the Mx and BF2 gene, Ri chickens resistant and susceptible to AIV were distinguished. The Mx protein, which is part a of the dynamin family of large GTPases, interferes with the replication of RNA viruses by inhibiting trafficking or the activity of viral polymerases [16–18]. The chicken major histocompatibility complex (MHC) consist of BLB (Class II) and two BF (Class I) [19, 20]. Especially, among haplotype of BF2, B21 allele is associated with high antibody titer against infectious bursal disease virus [21, 22]. Moreover, the chickens with B21 haplotype have high survival rate and with B13 have high mortality rate against H5N1 AIV [23]. To identify the profiling of exosomal miRNA against AIV infection, we infected Ri chickens with the AIV subtype H5N1 and purified exosomes from the serum for small RNA sequencing and analysis.
Materials and methods
Experimental birds and genotyping
All specifc-pathogen-free (SPF) Ri chickens [15], a native Vietnamese chicken breed, were infected with AIV and observed daily for signs of disease and mortality. All chicken experiments were performed in our collaborative laboratory at the Department of Biochemistry and Immunology in the National Institute of Veterinary Research, Vietnam. A total of 40 Ri chickens belonging to resistant and susceptible lines were used in our study (see Additional fle 1).
Genotyping of Mx and BF2 was performed by high resolution melt analysis for resistance and susceptibility selection [16–18, 22, 23]. In detail, the polymorphism of allele A/G of Mx at position 631 demonstrated allele A (resistant trait) and allele G (susceptible trait) in Ri chickens (see Additional file 2). Among BF2 haplotype, Ri chickens that have B21 haplotype were selected as a resistant trait and B13 haplotype were selected as a susceptible trait. Finally, Ri chickens Mx(A)/B21 were selected as a resistant trait and Mx(G)/B13 were selected as a susceptible trait. For HPAIV challenge, a total of 20 4-week-old birds, 10 Ri chickens in each line, received intranasal inoculation of 200 µL of harvested allantois fluid of the infected eggs, containing 1× 104 50% egg infectious dose (EID50) [24] of A/duck/Vietnam/ QB1207/2012 (H5N1), according to the OIE guideline.
Exosome extraction and characterization
Blood samples were collected from the wing vein of chickens after 1 and 3 days of infection (three chickens from each group). Exosomes were extracted from the serum using Total Exosome Isolation Reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. Briefly, 5 mL of the blood sample was collected from infected and control chickens. The blood was incubated at room temperature (RT) for 2 h to allow clotting. Serum was isolated from the clotted blood and centrifuged at 2000 × g for 30 min at 4 °C to remove cells and debris. The supernatant was mixed with 0.2 volumes of the Total Exosome Isolation reagent and incubated at 4 °C for 30 min. After incubation, samples were centrifuged at 10 000 × g for 10 min at RT. The supernatant was discarded, and exosomes were contained in the pellet at the bottom of the tube. Exosomes were suspended with phosphate-buffered saline (PBS; pH 7.4) and stored at ≤ −20 °C.
For characterization of exosomes, the particle size was measured using a Nanoparticle Analyzer (HORIBA, SZ-100, Kyoto, Japan). Furthermore, a Western blot assay was performed using CD81 as a exosomal marker (#56039; Cell Signaling Technology, Danvers, MA, USA) according to previously described methods [25].
Exosomal RNA extraction and small RNA sequencing
Small RNA sequencing was conducted using exosomes from resistant Ri chickens at day 3 post-infection. Exosomal RNA was extracted using the miRNeasy Serum/ Plasma Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Library construction and small RNA sequencing were only conducted for resistant Ri chickens with 1 control (Resistant day-3) and 2 infection (Resistant day-3) samples. The library was constructed using SMARTer smRNA-Seq Kit for Illumina (TAKARA Bio Inc., Otsu, Shiga, Japan) according to the manufacturer’s protocol. Next, small RNA sequencing was conducted by Macrogen (Seoul, Republic of Korea) using a HiSeq 2500 System (Illumina Inc., San Diego, CA, USA).
Bioinformatic analysis of sequencing data
The raw sequence reads were filtered based on quality using FastQC v0.11.7 [26]. Adapter sequences were trimmed of the raw sequence reads using Cutadapt 1.16 [27]. Both the trimmed and non-adapter reads were used as processed reads to analyze long targets (≥50 bp). Unique clustered reads were sequentially aligned to the reference genome using miRBase v21 [28], and the noncoding RNA database, RNAcentral 10.0 [29], to classify known miRNAs and other RNA types, such as tRNA, small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA). Novel miRNA prediction was performed by miRDeep2 [30]. The read counts for each miRNA were extracted from mapped miRNAs to report the abundance of each miRNA. Differentially expressed miRNAs were determined by comparing each miRNA across conditions using statistical methods, such as fold-change, exactTest using edgeR (Empirical Analysis of Digital Gene Expression Data in R), and hierarchical clustering. Target genes of differentially expressed miRNAs were predicted using miRDB [31] and target genes with a target score over 80 were selected. Next, GO functional enrichment analysis of target genes was performed using gProfiler [32]. The miRNA-mRNA network were constructed using miRNet [33].
miRNA primer design
Quantitative real-time PCR (qRT-PCR) for miRNAs only required a forward primer to be designed for the individual miRNA. The reverse primer was a universal primer provided with the miScript SYBR Green PCR Kit (Qiagen). All known chicken miRNA sequences were obtained from miRBase [34]. Oligonucleotide primers for these miRNAs were designed using full-length mature miRNA sequences. Primers were synthesized by Genotech (Daejeon, South Korea) (see Additional fle 3).
miRNA expression analysis by qRT‑PCR
For qRT-PCR, susceptible control (3 samples of day 3), susceptible infection (2 samples of day-3), resistant control (1 sample of day-3), and resistant infection (2 samples of day-3) RNA samples were used. cDNA synthesis was performed using miScript II RT Kit (Qiagen) according to the manufacturer’s protocol. Briefly, 1 μg of total RNA was combined with 4 μL of 5×miScript HiSpec Buffer, 2 μL of 10×miScript Nucleics Mix, 2 μL of miScript Reverse Transcriptase Mix, and RNase-free water up to 20 μL. The tube was incubated at 37 °C for 60 min and at 95 °C for 5 min to inactivate miScript Reverse Transcriptase Mix and kept on ice. Next, 20 μL of reverse-transcription reaction mixtures were diluted with 130 μL of RNase-free water. The synthesized cDNA was used as a template for qRT-PCR. miScript SYBR Green PCR Kit (Qiagen) was used to determine miRNA expression in LightCycler 96 system (Roche, Indianapolis, IN, USA) according to the manufacturers’ protocol. In brief, for 25 μL of reaction mix, the following components were added: 12.5 μL of 2×QuantiTect SYBR Green PCR Master Mix, 2.5 μL of 10 μM forward primer, 2.5 μL of 10×universal primer, 2.5 μL of template cDNA, and RNase-free water up to 25 μL. The cycling conditions were as follows: 95 °C for 15 min to activate as the initial step, followed by 45 cycles of 94 °C for 15 s, 55 °C for 30 s, and 70 °C for 30 s. Gene expression was calculated using the 2−ΔΔCt method after normalization with U1A (5′-CTGCATAATTTGTGGTAGTGG-3′) [35]. All qRTPCRs were performed in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS 25.0 software (IBM, Chicago, IL, USA). Data are expressed as mean values±SEM. Statistical comparisons were performed using Student’s t-test for two-group comparisons, and the level of statistically significant difference was set at p<0.05.
Results
Exosomal small RNA analysis
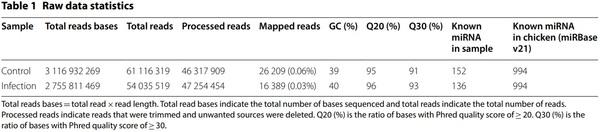
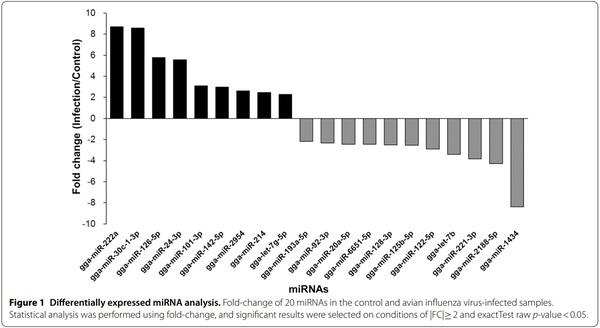
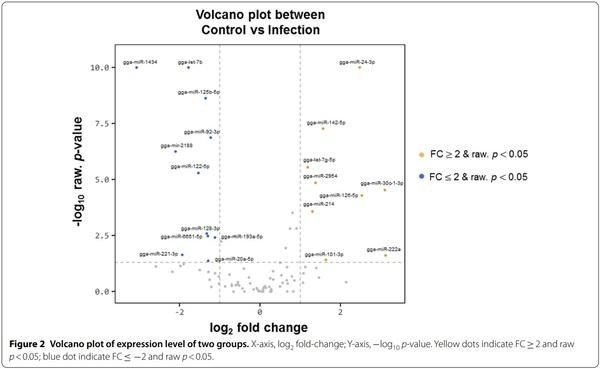
Exosomes were purified from the serum of resistant and susceptible Ri chickens at day 3 post-infection. The size and markers of exosomes were identified (see Additional file 4). Small RNA sequencing was conducted on resistant Ri chickens. In the control sample (Non-infected Ri chickens), 61 116 319 reads were produced, and total read bases were 3.1 Gbp (Table 1). In HPAIV-infected samples, 54 035 519 reads and 2.8 Gbp of read bases were produced. The GC content of control was 38.63% and the ratio of bases with Phred quality score ≥ 30 (Q30) was 90.99%. The GC content of infected chickens was 40.10% and the Q30 was 92.53%. Additional file 5 shows the read length distribution of each sample. In the control, a read length of approximately 17–20 bp was more abundant, whereas the read length was evenly distributed in infected chickens. For control and infected samples, final processed reads were sequentially aligned to the reference genome and the miRBase v21 and ncRNA databases to classify the known miRNAs and other types of RNA, such as tRNA, snoRNA, snRNA, and Piwi-interacting RNA (piRNA) (see Additional file 6). In the control, genome sequences are the largest part as a 92.56% and known miRNA, novel miRNA, snoRNA, snRNA, rRNA, tRNA account for 0.05%, 1.21%, 0.15%, 18.42%, 5.44%, and 0.03%, respectively. Also, genome sequences of the infected samples comprised the largest part (94.1%) and known miRNA, novel miRNA, snoRNA, snRNA, rRNA, and tRNA accounted for 0.03%, 0.32%, 0.04%, 6.46%, 2.25%, and 0.02%, respectively. To predict the known and novel miRNA, unique clustered reads were aligned against the reference genome and precursor miRNAs separately. Novel miRNAs are predicted from mature, star and loop sequence according to the RNAfold algorithm using miRDeep2. To detect known and novel miRNAs, miRDeep2 estimated their abundance (Table 1). In the control, among total 46 317 909 reads, 84.78% (39 269 083 reads) were mapped and 15.22% (7 048 826 reads) were unmapped reads. In infected chickens, among 47 254 454 reads, 90.21% (42 628 445 reads) were mapped and 9.79% (4 626 009 reads) were unmapped reads. We also investigated differentially expressed miRNA analysis by the read count value of mature miRNAs (see Additional file 7). A total of 20 miRNAs showed significantly different fold-change (Figure 1). Among 20 miRNAs, nine miRNAs were up-regulated and 11 miRNAs were down-regulated in infected chickens compared to those in the control. In particular, gga-miR-222a was the highest, with a fold-change of 8.69, and gga-miR-1434 was the lowest, with fold-change of −8.36. gga-miR-222a, gga-miR30c-1-3p, gga-miR-126-5p, gga-miR-24-3p, gga-miR101-3p, gga-miR-142-5p, gga-miR-2954, gga-miR-214, and gga-let-7g-5p were up-regulated in infected samples compared with that in the control, whereas gga-miR-193a-5p, gga-miR-92-3p, gga-miR-20a-5p, gga-miR-6651-5p, gga-miR-128-3p, gga-miR-125-5p, gga-miR-122-5p, gga-let-7b, gga-miR-221-3p, ggamiR-2188-5p, and gga-miR-1434 were down-regulated in infected samples compared with those in the control. We also compared the expression levels of miRNAs between control and infected samples using a volcano plot (Figure 2). Log2 fold-change and p-values obtained from the comparison between the two groups were plotted as a volcano plot. gga-miR-1434 displayed both high fold-change (x-axis) and statistical significance (y-axis). We also conducted hierarchical clustering analysis by the Euclidean method and complete linkage, which clusters similar mature miRNAs and samples by expression level (normalized value) from the differentially expressed miRNA list (Figure 3). Nine up-regulated exosomal miRNAs in the infected chickens were grouped in the generated dendrogram.
Target gene prediction, GO functional enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
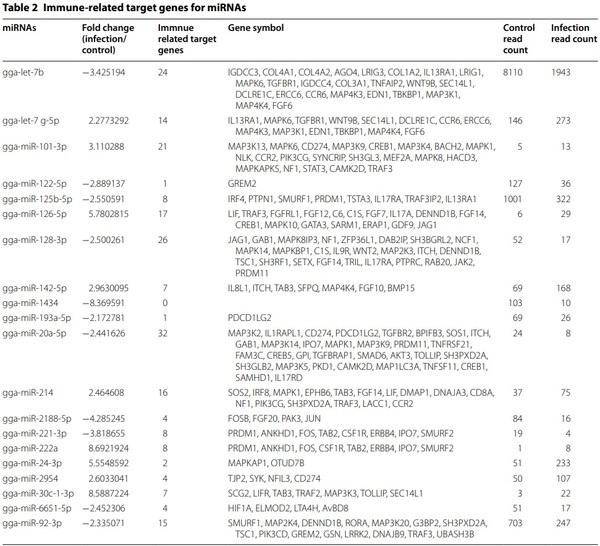
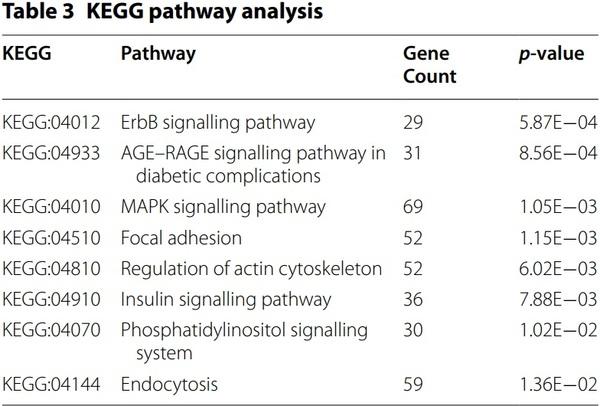
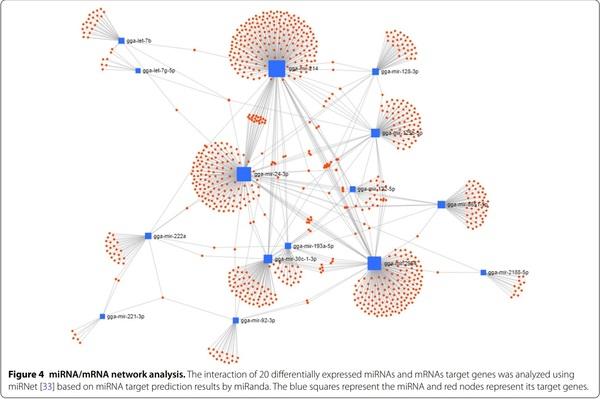
The target genes of 20 miRNAs were predicted using miRDB. Immune-related target genes with score of over 80 were selected (Table 2). In particular, 32 immune-related genes were related to gga-miR-20a-5p, and no immune-related target genes were related to ggamiR-1434. We also conducted GO analysis using gProfiler (see Additional file 8). In KEGG pathway analysis, eight pathways, including erythroblastic leukemia viral oncogene homolog (ErbB) signalling pathway, advanced glycation end-products (AGE)–receptor of AGE (RAGE) signalling pathway in diabetic complications, mitogenactivated protein kinase (MAPK) signalling pathway, focal adhesion, regulation of actin cytoskeleton, insulin signalling pathway, phosphatidylinositol signalling system, and endocytosis, were related to 20 miRNAs (Table 3). In particular, 69 target genes of miRNAs were related with the MAPK signalling pathway, followed by endocytosis (59 genes), focal adhesion (52 genes), and regulation of actin cytoskeleton (52 genes). Furthermore, the miRNA-mRNA target gene interaction of 20 differentially expressed miRNAs was analysed. As shown in Figure 4, several targets genes were shared with more than two miRNAs.
Validation of miRNA expression by qRT‑PCR
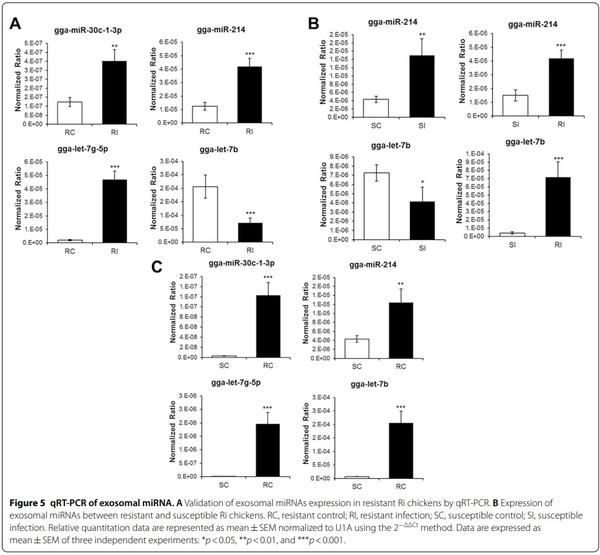
qRT-PCR was conducted using four miRNAs, selected on the basis of significant difference of>2.0 or<2.0 foldchange between control and infected chickens, to validate sequencing results (Figure 5A). The four miRNAs were selected based on read count, the number of immune-related genes, and functions in immune system. The expression levels of gga-miR-30c-1-3p, gga-miR-214, and gga-let-7g-5p were up-regulated in infected resistant chickens compared with those in the control by 2.85, 3.37, and 23.91 fold-change, respectively. However, the expression of gga-let-7b was down-regulated in infected chickens (0.34 fold-change). qRT-PCR results of the four miRNAs were positively correlated with sequencing results. The expression levels of gga-miR-214 and ggalet-7b were also evaluated in resistant and susceptible Ri chickens (Figure 5B). The expression level of gga-miR-214 was up-regulated in susceptible infected chickens compared with that in the control (3.48 fold-change). However, the expression level of gga-let-7b was downregulated in susceptible infected chickens compared with that in the control (0.57 fold-change). The expression patterns of gga-miR-214 and gga-let-7b were similar in susceptible and resistant chickens. Moreover, the expression levels of gga-miR-214 and gga-let-7b were higher (2.79 and 17.18 fold-change, respectively) in resistant chickens than in susceptible chickens. Furthermore, the expression of four miRNAs was compared between susceptible and resistant mock control (Figure 5C). The expression levels of gga-miR-214, gga-miR-30c-1-3p, gga-let-7g-5p, and gga-let-7b were higher in resistant than susceptible control chicken (2.88, 45.67, 180.39, and 28.34 fold-change, respectively).
Discussion
In this study, we report exosomal miRNA analysis of avian influenza virus infected chickens. The Vietnamese AIV-resistant Ri chickens were selected by Mx and BF2 genotyping and infected with H5N1 HPAIV. Then, exosomes were isolated from the serum, and miRNA was analysed by small RNA sequencing and qRT-PCR.
Exosomes are formed by various molecules, such as proteins, lipids, DNA, mRNA, and miRNA, that are contained in endosomes through endocytosis. Then, endosomes called MVBs fuse with the cell membrane and are released into the extracellular space [36]. Released exosomes containing miRNA are delivered to other cells through biological fluids and regulate the gene expression of recipient cells; however, they are not randomly packaged in exosomes [37–40].
In this study, 20 exosomal miRNAs exhibited significantly different expression between control and infected resistant Ri chickens (Figure 1). Previous studies have reported differentially expressed miRNA in chickens and humans. The expression of gga-miR-6651-5p increases in Marek’s disease virus infected-chickens [41]; gga-miR92-3p and gga-miR-214 are abundant in AIV H9N2- infected chicken embryo fibroblasts [42]; gga-miR-2954 is up-regulated in reticuloendotheliosis virus-infected chickens [43]; gga-miR-222a, gga-miR-125b-5p, and gga-miR-128-3p are down-regulated in H9N2-infected chicken embryo fibroblasts [42]. Likewise, in our study, gga-miR-125b and gga-miR-128-3p were down-regulated in H5N1-infected chickens. In another study, miR-126-5p inhibited human cervical cancer progression, thus, regulating the apoptosis of cancer cells [44]. gga-miR-101-3p is up-regulated in Mycoplasma gallisepticm-infected chickens [45]. gga-let-7b and miR-128 reduce cell growth and division during skeletal muscle development in sexlinked dwarf chickens [46]. gga-let-7b is involved in signalling pathways, such as MAPK, TGF-β, Notch, Wnt, mTOR, cell cycle, p53, and Janus-activated kinase (JAK)– signal transducers and activators of transcription (STAT) pathways [47]. Human miR-20a-5p inhibits cell proliferation and induces apoptosis in SH-SY5Y cells [48], and its down-regulation induces cell apoptosis to remove mycobacterial cells through targeting JNK2 in human macrophages [49]. Therefore, we suggest that 19 differentially expressed exosomal miRNAs from AI-infected chickens regulate immune response.
We analysed immune-related target genes (Table 2). Various immune-related genes, such as genes encoding signalling pathway molecules, cytokines, and chemokines, were found to be miRNA target genes. In particular, in KEGG pathway analysis, the highest number of target genes (69 gene count) were related with the MAPK signalling pathway (Table 3). The MAPK signalling pathway plays important roles in the immune system and also in cell proliferation, differentiation, migration, and apoptosis [50]. MAPK signalling pathway was activated by H5N1 AIV [51–53]. Several molecules of the MAPK pathway are differently regulated in two inbred necrotic enteritis-afflicted chicken lines, 6.3 and 7.2 [54]. Furthermore, the MAPK pathway plays an important role in virus replication in chicken macrophages infected with H9N2 AIV [55]. MAPK signalling pathway was followed by Endocytosis (59 gene count). As a first step of AIV infection, AIV enters the cells by endocytosis [56]. The next KEGG pathway was Focal adhesion and regulation of actin cytoskeleton (52 gene count). Focal adhesion kinase was activated during AIV infection by inducing actin rearrangement [57]. Therefore, we suggest that the target genes of 20 exosomal miRNAs regulate proinflammatory signalling pathway against AIV infection and life cycle of AIV.
We also compared the expression of gga-miR-214 and gga-let-7b between resistant and susceptible Ri chickens (Figure 4B). The expression patterns between control and infected chickens were the same, but the expression levels were higher in resistant than in susceptible chickens. Therefore, we suggest that the copy number of miRNAs is higher in the exosomes of resistant Ri chickens than in the exosomes of susceptible Ri chickens. Accordingly, regulation of gene expression by exosomal miRNAs may be higher in resistant Ri chickens. Hence, resistant Ri chickens could respond more actively than susceptible Ri chickens against AIV infection.
So far, several studies have analysed miRNA expression patterns on AIV infection [45, 58–61]. However, there have limited studies on the exosomes in chickens until now.
Taken together, this study provides an insight into the exosomal miRNA expression pattern in AIV-resistant and -susceptible chickens and the mechanism by which they regulate target genes toward HPAIV infection.
In summary, we have, for the first time, analysed the exosomal miRNA expression in H5N1 HPAIV-infected chickens by small RNA sequencing and qRT-PCR. A total of 20 miRNAs were differentially expressed in exosomes of control and infected resistant Ri chickens. Interestingly, most of the target genes were related with the MAPK signalling pathway. This study improves our understanding of the host immune response, particularly with respect to exosomal miRNA expression, against H5N1 HPAIV infection.















.jpg&w=3840&q=75)