Detection and Cellular Tropism of Porcine Astrovirus Type 3 on Breeding Farms
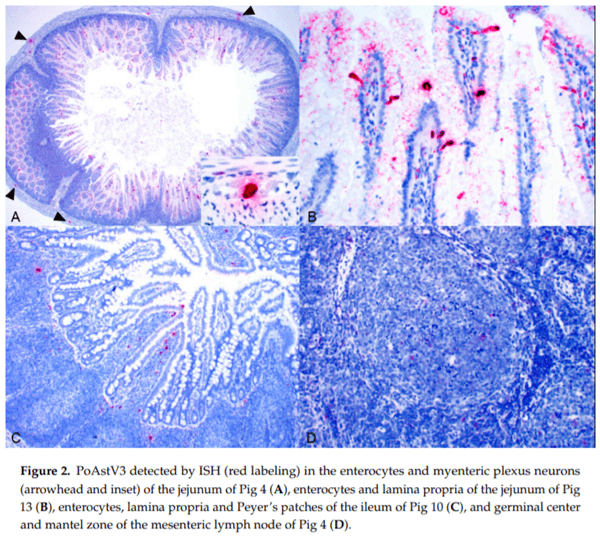
Astroviruses cause disease in a variety of species. Yet, little is known about the epidemiology of a majority of astroviruses including porcine astrovirus type 3 (PoAstV3), which is a putative cause of polioencephalomyelitis in swine. Accordingly, a cross-sectional study was conducted on sow farms with or without reported PoAstV3-associated neurologic disease in growing pigs weaned from those farms. Additionally, a conveniently selected subset of piglets from one farm was selected for gross and histologic evaluation. The distribution of PoAstV3 in the enteric system was evaluated through in situ hybridization. PoAstV3, as detected by RT-qPCR on fecal samples, was frequently detected across sows and piglets (66–90%) on all farms (65–85%). PoAstV3 was detected subsequently at a similar detection frequency (77% vs 85%) on one farm after three months. Viral shedding, as determined by the cycle quantification value, suggests that piglets shed higher quantities of virus than adult swine. No link between gastrointestinal disease and PoAstV3 was found. However, PoAstV3 was detected by in situ in myenteric plexus neurons of piglets elucidating a possible route of spread of the virus from the gastrointestinal tract to the central nervous system. These data suggest PoAstV3 has endemic potential, is shed in the feces at greater quantities by suckling piglets when compared to sows, and infection is widespread on farms in which it is detected.
Keywords: Porcine astrovirus type 3; astrovirus; detection; swine
1. Monroe, S.S.; Carter, M.J.; Herrmann, J.; Mitchell, J.R.; Sanchez-Fauquier, A. Astroviridae. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2005; pp. 859–864.
2. Bosch, A.; King, A.M.Q.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Family Astroviridae. Virus Taxonomy: Classification and Nomenclature of Viruses (Ninth Report of the International Committee on the Taxonomy of Viruses); Elsevier Academic Press: New York, NY, USA, 2011; pp. 953–959.
3. Bidin, M.; Lojki´c, I.; Tišliar, M.; Bidin, Z.; Majnari´c, D. Astroviruses associated with stunting and pre-hatching mortality in duck and goose embryos. Avian Pathol. 2012, 41, 91–97. [CrossRef] [PubMed]
4. Chu, D.K.; Poon, L.L.M.; Guan, Y.; Peiris, J.S.M. Novel astroviruses in insectivorous bats. J. Virol. 2008, 82, 9107–9114. [CrossRef] [PubMed]
5. Englund, L.; Chriél, M.; Dietz, H.H.; Hedlund, K.O. Astrovirus epidemiologically linked to pre-weaning diarrhoea in mink. Vet. Microbiol. 2002, 85, 1–11. [CrossRef]
6. Martella, V.; Moschidou, P.; Pinto, P.; Catella, C.; Desario, C.; Larocca, V.; Circella, E.; Bànyai, K.; Lavazza, A.; Magistrali, C.; et al. Astroviruses in rabbits. Emerg. Infect. Dis. 2011, 17, 2287–2293. [CrossRef] [PubMed]
7. Mor, S.K.; Abin, M.; Costa, G.; Durrani, A.; Jindal, N.; Goyal, S.M.; Patnayak, D.P. The role of type-2 turkey astrovirus in poult enteritis syndrome. Poult. Sci. 2011, 90, 2747–2752. [CrossRef] [PubMed]
8. Tse, H.; Chan, W.M.; Tsoi, H.W.; Fan, R.Y.; Lau, C.C.; Lau, S.K.; Woo, P.C.; Yuen, Y. Rediscovery and genomic characterization of bovine astroviruses. J. Gen. Virol. 2011, 92, 1888–1898. [CrossRef] [PubMed]
9. Xiao, C.T.; Giménez-Lirola, L.G.; Gerber, P.F.; Jiang, Y.H.; Halbur, P.G.; Opriessnig, T. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J. Gen. Virol. 2013, 94, 570–582. [CrossRef] [PubMed]
10. Benedictis, P.; Schultz-Cherry, S.; Burnham, A.; Cattoli, G. Astrovirus infections in humans and animals—Molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011, 11, 1529–1544. [CrossRef] [PubMed]
11. Luo, Z.; Roi, S.; Dastor, M.; Gallice, E.; Laurin, M.A.; L’Homme, Y. Multiple novel and prevalent astroviruses in pigs. Vet. Microbiol. 2011, 149, 316–323. [CrossRef] [PubMed]
12. Shan, T.; Li, L.; Simmonds, P.; Wang, C.; Moeser, A.; Delwart, E. The Fecal Virome of Pigs on a High-Density Farm. J. Virol. 2011, 85, 11697–11708. [CrossRef] [PubMed]
13. Lachapelle, V.; Letellier, A.; Fravalo, P.; Brassard, J.; L’Homme, Y. Dynamics of virus distribution in a defined swine production network using enteric viruses as molecular markers. Appl. Environ. Microbiol. 2017, 83, e03187-16. [CrossRef] [PubMed]
14. Cai, Y.; Yin, W.; Zhou, Y.; Li, B.; Ai, L.; Pan, M.; Guo, W. Molecular detection of Porcine astrovirus in Sichuan Province, China. Virol. J. 2016, 13, 6. [CrossRef] [PubMed]
15. Li, J.S.; Li, M.Z.; Zheng, L.S.; Liu, N.; Li, D.D.; Duan, Z.J. Identification and genetic characterization of two porcine astroviruses from domestic piglets in China. Arch. Virol. 2015, 160, 3079–3084. [CrossRef] [PubMed]
16. Xiao, C.T.; Luo, Z.; Lv, S.L.; Opriessnig, T.; Li, R.C.; Yu, X.L. Identification and characterization of multiple porcine astrovirus genotypes in Hunan province, China. Arch. Virol. 2017, 162, 943–952. [CrossRef] [PubMed]
17. Brni´c, D.; Prpi´c, J.; Keros, T.; Roi´c, B.; Starešina, V.; Jemerši´c, L. Porcine astrovirus viremia and high genetic variability in pigs on large holdings in Croatia. Infect. Genet. Evol. 2013, 14, 258–264. [CrossRef] [PubMed]
18. Dufkova, L.; S?cigalková, I.; Moutelíková, R.; Malenovská, H.; Prodelaova, J. Genetic diversity of porcine sapoviruses, kobuviruses, and astroviruses in asymptomatic pigs: An emerging new sapovirus GIII genotype. Arch. Virol. 2013, 158, 549–558. [CrossRef] [PubMed]
19. Amimo, J.O.; Okoth, E.; Junga, J.O.; Ogara, W.O.; Njahira, M.N.; Wang, Q.; Vlasova, A.N.; Saif, L.J.; Djikeng, A. Molecular detection and genetic characterization of kobuviruses and astroviruses in asymptomatic local pigs in East Africa. Arch. Virol. 2014, 159, 1313–1319. [CrossRef] [PubMed]
20. Goecke, N.B.; Hijulager, C.K.; Kongsted, H.; Boye, M.; Rasmussen, S.; Granberg, F.; Fischer, T.K.; Midgley, S.E.; Rasmussen, L.D.; Angen, Ø.; et al. No evidence of enteric viral involvement in the new neonatal porcine diarrhoea syndrome in Danish pigs. BMC Vet. Res. 2017, 13, 315. [CrossRef] [PubMed]
21. Machnowska, P.; Ellerbroek, L.; Johne, R. Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Vet. Microbiol. 2014, 168, 60–68. [CrossRef] [PubMed]
22. Sachsenröeder, J.; Twardziok, S.; Hammerl, J.A.; Janczyk, P.; Wrede, P.; Hertwig, S.; Johne, R. Simultaneous Identification of DNA and RNA Viruses Present in Pig Faeces Using Process-Controlled Deep Sequencing. PLoS ONE 2012, 7, e34631. [CrossRef] [PubMed]
23. Boros, A.; Albert, M.; Pankovics, P.; Bíró, H.; Pesavento, P.A.; Phan, T.G.; Delwart, E.; Reuter, G. Outbreaks of Neuroinvasive Astrovirus Associated with Encephalomyelitis, Weakness, and Paralysis among Weaned Pigs, Hungary. Emerg. Infect. Dis. 2017, 23, 1999–2010. [CrossRef] [PubMed]
24. Monini, M.; Di Bartolo, I.; Ianiro, G.; Angeloni, G.; Magistrali, C.F.; Ostanello, F.; Ruggeri, F.M. Detection and molecular characterization of zoonotic viruses in swine fecal samples in Italian pig herds. Arch. Virol. 2015, 160, 2547–2556. [CrossRef] [PubMed]
25. Ito, M.; Kuroda, M.; Masuda, T.; Akagami, M.; Haga, K.; Tsuchiaka, S.; Kishimoto, M.; Naoi, Y.; Sano, K.; Omatsu, T.; et al. Whole genome analysis of porcine astroviruses detected in Japanese pigs reveals genetic diversity and possible intra-genotypic recombination. Infect. Genet. Evol. 2017, 50, 38–48. [CrossRef] [PubMed]
26. Salamunova, S.; Jackova, A.; Mandelik, R.; Novotny, J.; Vlasakova, M.; Vilcek, S. Molecular detection of enteric viruses and the genetic characterization of porcine astroviruses and sapoviruses in domestic pigs from Slovakian farms. BMC Vet. Res. 2018, 14, 313. [CrossRef] [PubMed]
27. Blomstrom, A.L.; Cecilia, L.; Jacobson, M. Astrovirus as a possible cause of congenital tremor type AII in piglets? Acta Vet. Scand. 2014, 56, 82. [CrossRef] [PubMed]
28. Karlsson, O.E.; Larsson, J.; Hayer, J.; Berg, M.; Jacobson, M. The Intestinal Eukaryotic Virome in Healthy and Diarrhoeic Neonatal Piglets. PLoS ONE 2016, 11, e0151481. [CrossRef] [PubMed]
29. Lee, M.H.; Jeoung, H.Y.; Park, H.R.; Lim, J.A.; Song, J.Y.; An, D.J. Phylogenetic analysis of porcine astrovirus in domestic pigs and wild boars in South Korea. Virus Genes 2013, 46, 175–181. [CrossRef] [PubMed]
30. Kumthip, K.; Khamrin, P.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Ushijima, H.; Maneekarn, N. Detection and genetic characterization of porcine astroviruses in piglets with and without diarrhea in Thailand. Arch. Virol. 2018, 163, 1823–1829. [CrossRef] [PubMed]
31. Arruda, B.; Arruda, P.; Hensch, M.; Chen, Q.; Zheng, Y.; Yang, C.; Gatto, I.R.H.; Matias, F.F.; Gauger, P.; Schwartz, K.; et al. Porcine Astrovirus Type 3 in Central Nervous System of Swine with Polioencephalomyelitis. Emerg. Infect. Dis. 2017, 23, 2097–2100. [CrossRef] [PubMed]
32. Matias, F.F.; Arruda, P.; Hench, M.; Gatto, I.R.H.; Hause, B.; Li, G.; Chen, Q.; Zheng, Y.; Yang, C.; Harmon, K.; et al. Identification of a divergent porcine astrovirus type 3 in central nervous system tissue from swine with neurologic disease and encephalomyelitis: Diagnostic investigation, virus characterization and retrospective analysis of historic cases. In Proceedings of the American Association of Veterinary Laboratory Diagnosticians (AAVLD) Annual Conference Proceedings, San Diego, CA, USA, 12–18 October 2017.
33. Mor, S.K.; Chander, Y.; Marthaler, D.; Patnayak, D.P.; Goyal, S.M. Detection and molecular characterization of Porcine astrovirus strains associated with swine diarrhea. J. Vet. Diagn. Investig. 2012, 24, 1064–1067. [CrossRef] [PubMed]
34. National Hog Farmer. Porcine Astrovirus Type 3 Emerging Cause of Neurologic Disease in Swine. Available online: https://www.nationalhogfarmer.com/animal-health/porcine-astrovirus-type-3-emergingcause-neurologic-disease-swine (accessed on 15 August 2019).
35. Rawal, G.; Matias, F.F.; Mueller, A.; Allison, G.; Hedberg, W.; Macedo, N.; Bradner, L.; Harmon, K.; Linhares, D.; Arruda, B. Porcine astrovirus type 3 is an emerging cause of atypical neurologic disease: Diagnostic cases and infection dynamics on affected flows. In Proceedings of the 50th Annual Meeting of the American Association of Swine Veterinarians (AASV), Orlando, FL, USA, 9–12 March 2019.
36. Matias, F.F.; Bradner, L.K.; Burrough, E.R.; Cooper, V.L.; Derscheid, R.J.; Gauger, P.C.; Harmon, K.M.; Madson, D.; Piñeyro, P.E.; Schwartz, K.J.; et al. Polioencephalomyelitis in Domestic Swine Associated with Porcine Astrovirus Type 3. Vet. Pathol. 2019. [CrossRef]
37. Schroeder, M.E.; Bounpheng, M.A.; Rodgers, S.; Baker, R.J.; Black, W.; Naikare, H.; Velayudhan, B.; Sneed, L.; Szonyi, B.; Clavijo, A. Development and performance evaluation of calf diarrhea pathogen nucleic acid purification and detection workflow. J. Vet. Diagn. Investig. 2012, 24, 945–953. [CrossRef] [PubMed]
38. Cordey, S.; Vu, D.L.; Schibler, M.; L’Huilier, A.G.; Brito, F.; Docquier, M.; Posfay-Barbe, K.M.; Petty, T.J.; Turin, L.; Zdobnov, E.M.; et al. Astrovirus MLB2, a New Gastroenteric Virus Associated with Meningitis and Disseminated Infection. Emerg. Infect. Dis. 2016, 22, 846–853. [CrossRef] [PubMed]
39. Li, L.; Diab, S.; McGraw, S.; Barr, B.; Traslavina, R.; Higgins, R.; Talbot, T.; Blanchard, P.; Rimoldi, G.; Fahsbender, E.; et al. Divergent astrovirus associated with neurologic disease in cattle. Emerg. Infect. Dis. 2013, 19, 1385–1392. [CrossRef] [PubMed]
40. Pfaff, F.; Schlottau, K.; Scholes, S.; Courtnenay, A.; Hoffmann, B.; Höper, D.; Beer, M. A novel astrovirus associated with encephalitis and ganglionitis in domestic sheep. Transbound. Emerg. Dis. 2017, 64, 677–682. [CrossRef] [PubMed]
41. Quan, P.L.; Wagner, T.A.; Briese, T.; Torgerson, T.R.; Hornig, M.; Tashmukhamedova, A.; Firth, C.; Palacios, G.; Baisre-De-Leon, A.; Paddock, C.D.; et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 2010, 16, 918–925. [CrossRef] [PubMed]
42. Virus Survival in the Environment with Special Attention to Survival in Sewage Droplets and other Environmental Media of Fecal or Respiratory Origin. Available online: http://www.oretek.com/drugs/WHO_ VirusSurvivalReport_21Aug2003.pdf (accessed on 15 August 2019).
43. Shimizu, M.; Shirai, J.; Narita, M.; Yamane, T. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J. Clin. Microbiol. 1990, 28, 201–206. [PubMed]
44. Abad, F.X.; Pintó, R.M.; Villena, C.; Gajardo, R.; Bosch, A. Astrovirus survival in drinking water. Appl. Environ. Microbiol. 1997, 63, 3119–3122. [PubMed]
45. Banos-Lara, M.R.; Mèndez, E. Role of individual caspases induced by astrovirus on the processing of its structural protein and its release from the cell through a non-lytic mechanism. Virol. J. 2010, 401, 322–332. [CrossRef] [PubMed]
46. Arias, C.F.; DuBois, M. The Astrovirus Capsid: A Review. Viruses 2017, 9, 15. [CrossRef] [PubMed]
47. Curtis, R.; Adryan, K.M.; Zhu, Y.; Harkness, P.J.; Lindsay, R.M.; DiStefano, P.S. Retrograde axonal transport of ciliary neurotrophic factor is increased by peripheral nerve injury. Nature 1993, 365, 253–255. [CrossRef] [PubMed]
48. Gromeier, M.; Wimmer, E. Mechanism of injury-provoked poliomyelitis. J. Virol. 1998, 72, 5056–5060. [PubMed]
49. Ren, R.; Racaniello, V.R. Poliovirus spreads from muscle to the central nervous system by neural pathways. J. Infect. Dis. 1992, 166, 747–752. [CrossRef] [PubMed]
50. Lancaster, K.Z.; Pfeiffer, J.K. Limited Trafficking of a Neurotropic Virus Through Inefficient Retrograde Axonal Transport and the Type I Interferon Response. PLoS Pathog. 2010, 6, e1000791. [CrossRef] [PubMed]






