Introduction
Feed manufacturing equipment has been shown to be a potential source of porcine epidemic diarrhea virus (PEDV) cross contamination. Wet decontamination has been found to be the most effective method for decontaminating the surface of feed mill equipment. However, this is not practical in most current commercial feed production settings. Methods to mitigate the risk of PEDV transmission in feed and feed ingredients have been investigated, including chemical mitigation using products such as formaldehyde, medium chain fatty acids, essential oils, or dietary acidifiers, as well as thermal mitigation accomplished by pelleting diets. These methods are not universally applicable to all feed manufacturing facilities due to equipment cost and/or lack of application equipment.
Another key deterrent from using chemical mitigation strategies in complete swine diets is the economic burden from including products in all feed that is produced. Other research has assessed sequencing batches of PEDV-negative feed following an inoculated batch of feed to assess the effectiveness of reducing the risk of viral transmission. A series of diets intended for low-risk cohorts of livestock would be manufactured to essentially “flush” the system prior to manufacturing diets intended for high-risk groups, such as breeding stock. While this may be the most practical mitigation technique for feed mills to implement, there still remains a significant quantity of viral particles on feed-contact surfaces, and environmental contamination, including dust production and distribution throughout the facility. This dust may pose a risk for contamination of subsequent diets. One potential solution is to use chemical mitigants as a periodic flush step within the feed manufacturing process. The periodic nature would reduce the cost and may be an easily implementable, more cost-effective mitigation strategy than using chemical mitigants in each batch. Rice hulls were selected as the carrier for this chemical flush because the ingredient is commonly used as a carrier for vitamins and other micronutrients due to its relatively low cost. Furthermore, it has a high degree of abrasiveness, which may help facilitate the removal of viral contamination on equipment surfaces. Therefore, the objective of this experiment was to determine the impact of MCFA- or formaldehyde-treated rice hull flush batches as potential PEDV mitigation strategies during feed manufacturing.
Procedures
General
The experiment was conducted at the Kansas State University Cargill Feed Safety Research Center (FSRC). Prior to the experiment, the FSRC was decontaminated following a standard protocol approved by the Kansas State University Institutional Biosafety Committee. Prior to initiation of the experiment, the FSRC was physically cleaned using compressed air and sweeping, chemically cleaned using a two-step process using a 1:256 dilution of ammonium glutaraldehyde blend (Synergize; Preserve International, Reno, NV) and a 1:32 dilution of sodium hypochlorite solution. The facility was then heated to 140°F for a minimum of 24 h and cooled to room temperature at which point the environmental surfaces were sampled (World Bioproducts, Mundelein, IL) and verified devoid of PEDV viral RNA to ensure efficacy of the disinfection procedures prior to initiation of the experiment. After chemical disinfection, the facility was held in containment mode with negative air pressure and High-Efficiency Particulate Arrestance (HEPA) filters preventing contaminated air from leaving the facility. Containment was maintained throughout the experiment and through the post-decontamination procedures using the same procedures described above.
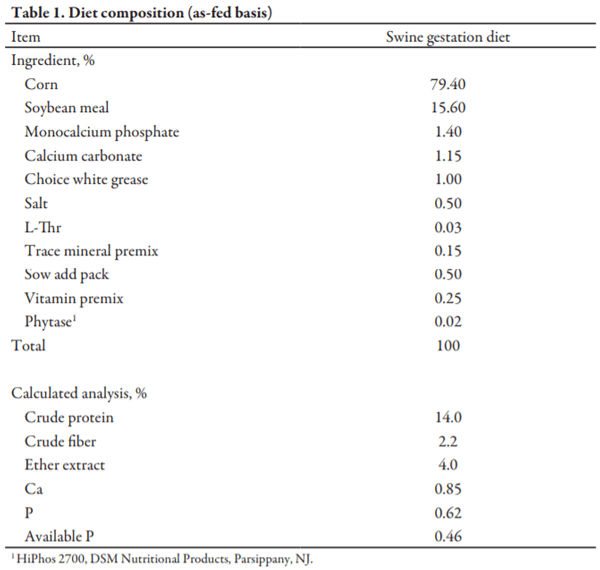
The swine diet (Table 1) used in this experiment was manufactured at the Kansas State University O.H. Kruse Feed Technology Innovation Center in Manhattan, KS, and was verified to be devoid of PEDV and porcine delta-coronavirus (PDCoV), viral RNA as determined via qRT-PCR prior to initiation of the experiment. Rice hulls were also analyzed using qRT-PCR prior to the experiment to confirm lack of PEDV genetic material. The production-scale mixer used was a 0.14-yd electric paddle mixer (H.C. Davis Sons Manufacturing, model # SS-L1; Bonner Springs, KS) with a mix time of 5 min. Feed was discharged at a rate of approximately 10 lb/min into a bucket elevator (Universal Industries, Cedar Falls, IA) fitted with 74 buckets (114 cm3 each), and then discharged through a 10 inch diameter discharge spout, and collected in plastic biohazard bags. Laboratory-scale stainless steel paddle mixers (n = 13; Cabela’s Inc., Sidney, NE) were validated for mixer efficiency for 5.5 and 11.0 lb batches using a mix time of 5 min. Validation of mixers prior to the experiment to achieve a coefficient of variation of less than 10% was done following previously described procedures.
Chemical Treatment of Flush Batches and Negative Samples
Prior to initiation of the experiment, six 5.5 lb chemically-treated rice hull batches were prepared using 2% MCFA blend (n = 2; 1:1:1 ratio of caproic, caprylic, and capric acid), 10% MCFA blend (n = 2; same ratio of acids used as in 2% blend), or commercial formaldehyde (n = 2; Sal CURB, Kemin Industries, Inc.; application rate = 6.5 lb/T). Untreated rice hulls (5.5 lb; n = 2) were also weighed and prepared prior to initiation of the experiment. Rice hulls (untreated and chemically-treated) were stored in double lined bags for 48 hours at room temperature (70°F) until initiation of experiment.
Prior to inoculation with PEDV, batches of feed were placed in, mixed, and discharged through both a laboratory-scale mixer and production-scale systems. For the laboratory-scale mixers, 500 g of PEDV negative feed was added to each mixer, rotated for approximately 15 sec, then disconnected from the drive unit and inverted in a one-step motion to dispose of feed into a waste container. A small quantity of residual feed remained in each mixer after this systematic priming discharge procedure. Following the priming of each mixer, a 5.5 lb batch of PEDV-negative feed was added to each laboratory-scale mixer and mixed as described above. The mixer then was shut off, drive coupler removed from the drive unit motor, and a subsample was collected from six locations within each mixer for a total sample size of approximately 0.5 lb. The mixer was then fully disconnected and inverted to dispose of feed into a waste container.
After priming and collection of the negative feed sample from laboratory-scale mixer, the production-scale system was primed and negative sample collected. An 11 lb batch of PEDV-negative feed was added to the production-scale mixer, allowed to mix for approximately 15 seconds, and subsequently discharged into the bucket elevator and was collected at the discharge spout to prime the mixer and fill the boot of the bucket elevator. A 110 lb batch of PEDV-negative feed was then added to the production-scale mixer, mixed for 5 min, and then discharged into the bucket elevator and collected in bags at the discharge spout. A sample of feed was collected from multiple subsample points within the discharged batch of feed.
Laboratory-Scale Mixer Inoculation, Flush, and Subsequent Feed
The viral inoculum was cell culture derived (USA/IN/2013/19338, passage 9) and had an initial concentration of 4 × 106 TCID50/mL. A 1:10 dilution was performed using phosphate buffered saline (PBS; pH 7.4 1X, Life Technologies, Grand Island, NY) to create 2,500 mL of 105 TCID50/mL viral inoculum. Inoculation of feed to be used in each of the laboratory-scale mixers was performed in 11.0 lb batches using an additional laboratory-scale mixer in which 9.9 lb of PEDV negative feed was added to the mixer and 500 mL of 105 TCID50/mL diluted viral inoculum was added to create 11.0 lb of 104 TCID50/g inoculated feed. This batch was mixed for 5 min, at which point it was split into two samples using a riffle splitter and weighed into 5.5 lb batches, bagged, and stored in a freezer (10°F) until inoculated into appropriate laboratory-scale mixer. This process was repeated three additional times, to create a total of eight 5.5 lb batches of inoculated feed.
After preparation of laboratory-scale mixer inoculated feed, each of 8 laboratory-scale mixers was inoculated with feed, flush step performed, and a subsequent batch of feed was mixed and sampled. For each inoculation, a bagged sample of PEDV-inoculated feed was randomly selected from the freezer and placed into the randomly selected laboratory-scale mixer. Feed was mixed for 5 min, at which point a sample of PEDVinoculated feed was collected from 6 locations within the mixer. PEDV-inoculated feed was then discarded into biohazard waste bags using a complete inversion of the mixer following the systematic procedure as described above with no tapping or additional cleaning action. The appropriate flush batch was added to the mixer and mixed for 5 min. A sample of the rice hull flush was collected from 6 locations within the mixer as previously described. The remaining flush was then discarded, and a subsequent 5.5 lb batch of PEDV-negative feed was added to the mixer and mixed. After mixing, a sample of the subsequent feed was collected, and remaining feed was discarded. This process was repeated 7 additional times in a random order blocked by repetition number, for a total 8 laboratory-scale mixers with two replicates of each of the four chemical treatments (untreated rice hulls, Sal CURB-treated rice hulls, 2% MCFA-treated rice hulls, and 10% MCFA-treated rice hulls).
Production-Scale System Inoculation, Flush, and Subsequent Feed
For inoculation of the production-scale system, a 9.9 lb batch of PEDV-negative feed was added to a clean laboratory-scale paddle mixer and 500 mL of 106 TCID50/mL inoculum was slowly added to create an 11 lb batch of PEDV inoculated feed (105 TCID50/g). Upon conclusion of the addition of the virus, the batch was mixed for 5 min to ensure an even mix of virus into the feed inoculum. The PEDV feed inoculum was then added to 99 lb of PEDV-free swine diet in the production-scale mixer to create the 110-lb batch of PEDV positive feed (104 TCID50/g). The entire batch of PEDV-positive feed was then mixed for 5 min, discharged into the bucket elevator, and collected at the bucket elevator discharge spout in biohazard waste bags. A sample of PEDV-positive feed was collected from multiple locations within the discharged batch of PEDV-positive feed. This sample of PEDV inoculated feed was combined at a 1:1 ratio with PEDV-inoculated feed (also 104 TCID50/g) from laboratory-scale mixer to create a single PEDV-positive sample.
After inoculation of the production-scale mixer, 79.2 lb of ground rice hulls were added directly to the mixer, along with 8.8 lb of MCFA (1:1:1 ratio of caproic, caprylic, and capric acid) to create a 10% MCFA rice hull flush with a similar mixer fill volume as a 110 lb batch of feed. After a 5 min mix time, 6 samples were collected from various locations within the mixer. The rice hull flush batch was then discharged into the bucket elevator and collected at the bucket elevator discharge spout. Samples of discharged flush material were collected at multiple times during discharge to create a single composite sample.
A 110 lb batch of PEDV-negative feed was then added to the production-scale mixer and allowed to mix for 5 min. A 0.5 lb sample was collected from the mixer and remaining feed was discharged into the bucket elevator and collected at the bucket elevator discharge spout. Another 0.5 lb sample was collected from six locations of the bucket elevator to create a single composite sample. Samples were placed on ice and transported to the laboratory for qRT-PCR analysis preparation.
Dust samples were also collected throughout the experiment, including dust collected after mixing of 104 TCID50/g inoculated feed in both the laboratory and production-scale systems, after mixing of 10% MCFA treated rice hulls in the production-scale mixer, and collected from mixing of the subsequent feed following the 10% MCFA rice hull flush. All dust collection surfaces were above the fill level of the mixer; therefore, all collected dust had become airborne before depositing on the collection surfaces. Dust was collected from the same surface after each batch of feed (positive inoculated feed, 10% MCFA rice hull flush, and subsequent PEDV-free feed); therefore, dust collected was produced during the associated mixing process and not from previous manufacturing processes.
Viral RNA Quantification
After sample collection, temporary storage on ice, and transport to the Kansas State University Molecular Diagnostic Research and Development Laboratory, three 50.0 g subsamples of feed from each collection point were added to individual 500 mL high density polyurethane (HDPE) bottles. Rice hull samples from each collection point were subsampled into three 25.0 g samples and added to individual 250 mL HDPE bottles. After subsampling of all feed and rice hull flush samples into appropriate bottles, varying quantities of PBS (100 or 200 mL for rice hull or feed, respectively) were added to each bottle to create a 20% suspension. Bottles were shaken for approximately 10 sec, at which point they were allowed to settle overnight at 39.2°F. On the next day, supernatant was collected and multiple aliquots prepared for further analysis. A total of 4 aliquots from each sample bottle were collected and stored at -4°F until qRT-PCR analysis was performed within 7 d of inoculation on one aliquot per sample bottle. The remaining three samples per bottle were stored at -112°F until further use. Dust samples were subsampled into 1 mL aliquots, and 4 mL of PBS was added resulting in a 20% suspension by volume. Samples were stored in a similar manner to feed and rice hull flush bottles, and supernatant pulled the following day to be analyzed via qRT-PCR. The remaining dust was stored in dry form at -112°F until initiation of the bioassay portion of the experiment.
Polymerase chain reaction (PCR) assays were performed at the Kansas State University Molecular Diagnostic Research and Development Laboratory. Briefly, fifty microliters (µL) of supernatant from each sample were loaded into a deep well plate and extracted using a Kingfisher 96 magnetic particle processor (Fisher Scientific, Pittsburg, PA) and the MagMAX-96 Viral RNA Isolation kit (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions with one modification, reducing the final elution volume to 60 µL. One negative extraction control consisting of all reagents except the sample was included in each extraction. The extracted RNA was frozen at -4°F until assayed by qRT-PCR. Reported values represent threshold cycle time (Ct) at which virus was detected. A greater Ct value indicates more cycles must proceed until viral genetic material was detected, thus representing lower quantities of genetic material in the original sample.
Infectivity Characteristics
Bioassay samples were selected after qRT-PCR analysis, and included a composite positive and negative control sample that had been collected from both laboratory and production-scale mixers, untreated rice hull flush, 2% MCFA rice hull flush, Sal CURB rice hull flush, subsequent feed after the untreated rice hull flush, subsequent feed after Sal CURB rice hull flush, subsequent feed after the 2% MCFA rice hull flush, subsequent feed after the 10% MCFA rice hull flush, 10% MCFA rice hull flush collected from the discharge spout of the production-scale system, and subsequent feed after the 10% MCFA rice hull collected from the discharge spout of the production-scale system. Additionally, dust samples included in the bioassay were those collected from mixing surfaces after manufacture of 104 TCID50/g inoculated swine feed, dust collected after the 10% MCFA rice hull flush, and subsequent feed after the 10% MCFA rice hull flush. Supernatant samples were allowed to thaw prior to bioassay inoculation at room temperature, beginning approximately 3 h prior to inoculation. Dust samples were prepared by combining the three positive control dust samples into a single, homogeneous positive control dust sample. A total of three, homogeneous, dust samples (positive, 10% MCFA rice hull flush, and subsequent feed dust) were then each split into three 5.2 g aliquots, and then adding 20.8 g PBS to create a 1:5 suspension of dust to total mass, with a volume of approximately 25 mL each. A 1 mL sample of the suspension was sampled for qRT-PCR analysis, and the remaining solution was inoculated into the appropriate pig (n = 3 pigs per dust type).
The experimental protocol for the pig bioassay portion of the experiment was reviewed and approved by the Iowa State University Institutional Animal Care and Use Committee. Forty-two crossbred, 10 d-old pigs of mixed gender were sourced from a single commercial, crossbred farrow-to-wean herd with no known prior exposure to PEDV. Pigs received a dose of cefitiofur (Excede, Zoetis, Florham Park, NJ) just prior to transport to the research facility. Upon arrival, piglets were ear tagged, weighed and randomly assigned to bioassay treatment rooms. Fecal swabs were obtained and confirmed negative for PEDV, porcine delta coronavirus (PDCoV) and transmissible gastroenteritis virus (TGEV) using qRT-PCR analysis. To further confirm PEDV negative status, serum was collected and confirmed negative for PEDV antibody by an indirect fluorescent antibody (IFA) assay and TGEV antibody by ELISA conducted at the Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL). Pigs were allowed 2 d of adjustment to the new pens before inoculation.
Pigs from each treatment were housed in separate rooms with independent ventilation systems. Rooms had solid flooring that was minimally rinsed to reduce risk of PEDV aerosols. Pigs were fed liquid milk replacer once daily and offered a commercial pelleted swine diet ad libitum with free access to water. Each of 33 pigs (11 rooms) receiving supernatant samples were administered 20 mL of the PBS supernatant by orogastric gavage using a 10-gauge French catheter 0 days post-bioassay inoculation (dpi). Each of 9 pigs (3 rooms) which were inoculated with dust suspensions by administering the separated supernatant via orogastric gavage (approximately 5 to 10 mL), with the remaining solid fraction of the inoculum directly placed in the mouth of each pig at which point they were stimulated to swallow. Rectal swabs were collected daily throughout the bioassay experiment from all piglets, and tested for PEDV RNA via qRT-PCR on d -2, 0, 2, 4, 6, and 7 dpi. Fresh small intestine, cecum, and colon were collected at necropsy at 7 dpi, along with an aliquot of cecal content. One section of formalin-fixed proximal, middle, and distal jejunum and ileum were collected for histopathology. Cecal content was evaluated for presence of PEDV genetic material via qRT-PCR. Tissue was processed and fixed in neutral buffered formalin, embedded, sectioned, and stained with hematoxylin and eosin stain. One section of proximal, middle, and distal jejunum; and three serial sections from the piece of ileum (for a total of six sections of intestine) were evaluated by a veterinary pathologist blind to the treatments.
Statistical Analysis
Data were analyzed using PROC GLIMMIX (SAS Institute, Inc., Cary, NC) to determine differences between the treatments. Pairwise comparisons were used to determine differences among flush strategies, with the model protected by the overall F-test. Bottle was included in the model as a random effect. A cycle time value of 45 was used in the statistical analysis for samples not containing detectable genetic material. Results for response criteria were considered significant at P ≤ 0.05.
Results and Discussion
Viral RNA Quantification
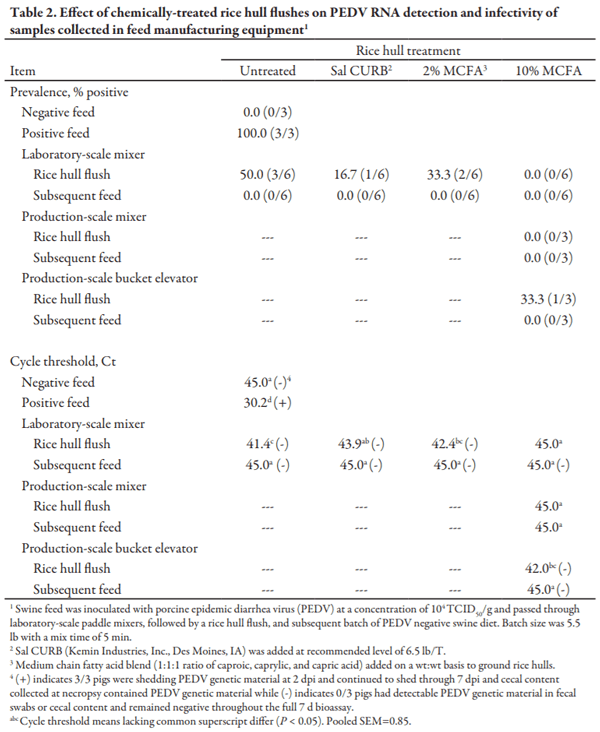
After qRT-PCR analysis, the composite negative feed sample did not have detectable RNA, and composite positive control feed sample contained detectable PEDV genetic material (Table 2). Following a PEDV positive batch of feed in laboratory-scale mixers, 50% of the untreated rice hull flush samples had detectable PEDV RNA. The untreated rice hull flush reduced (P < 0.05) the quantity of detectable RNA compared to the PEDV positive batch of feed. One Sal CURB treated rice hull flush sample was positive for PEDV genetic material, and 33% of the 2% MCFA rice hull samples had detectable PEDV RNA. Additionally, none of the 10% MCFA rice hull flush samples had detectable PEDV RNA. Chemically-treated rice hull flushes using Sal CURB and 10% MCFA reduced (P < 0.05) the quantity of detectable RNA present in the rice hull flush samples compared to the untreated rice hull flush. However, the 2% MCFA rice hull flush did not reduce (P = 0.215) the quantity of genetic material compared to the untreated rice hull flush. Importantly, no feed samples after an untreated or chemically-treated rice hull flush had detectable PEDV RNA.
After manufacturing a PEDV-positive batch of feed in the production-scale mixer and bucket elevator, one 10% MCFA rice hull sample collected from the bucket elevator discharge spout had detectable RNA. However, none of the rice hull flush samples collected from the mixer or subsequent feed samples from the mixer or bucket elevator discharge spout had detectable PEDV RNA. The presence of detectable viral RNA in the 10% MCFA-treated rice hull flush sample collected from the bucket elevator discharge spout provides evidence that bucket elevators are a significant source of cross contamination within feed manufacturing systems.
All pigs were free of PEDV genetic material in fecal swabs and PEDV-specific antibodies prior to initiation of the bioassay experiment. On 2 dpi, fecal shedding of PEDV RNA was detected in positive control pigs. No other flush feed bioassay pigs had detectable RNA in fecal swabs throughout the study or cecal content collected at necropsy.
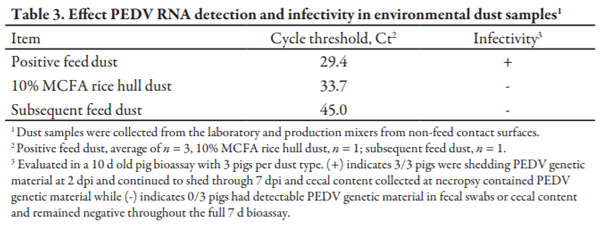
Dust collected after mixing the positive feed had a large quantity of viral RNA (Table 3). Following the inoculated batch of feed, dust collected immediately following the 10% MCFA rice hull flush batch had a reduced quantity of viral RNA, and subsequent feed following the 10% MCFA rice hull flush did not have detectable RNA. Pigs inoculated with the positive dust collected following mixing of inoculated feed were shedding PEDV by d 2 after oral inoculation and continued to shed through necropsy at 7 dpi. However, pigs inoculated with the dust from the 10% MCFA rice hull flush batches or the subsequent feed batch had no indications of PEDV infection.
Overall, the rice hull flush effectively reduced the amount of detectable RNA present compared to feed inoculated with PEDV, as expected. Chemical treatment of rice hulls with Sal CURB and 10% MCFA provided additional reduction in detectable RNA present in the rice hull flush samples. Feed manufactured following rice hull flushes did not contain PEDV RNA, therefore utilizing rice hull flushes would be a cost effective strategy to mitigate the risk of PEDV transmission via the feed manufacturing process. Finally, dust collected following the manufacture of PEDV inoculated feed contained a large quantity of PEDV RNA and was infective. Therefore, a high level of caution should be taken to limit and control dust created during feed manufacturing as it may serve as a vector in PEDV transmission.
This article was originally published in Kansas Agricultural Experiment Station Research Reports: Vol. 2: Iss. 8. https://doi.org/10.4148/2378-5977.1282. This is an Open Access article licensed under a Creative Commons Attribution 4.0 License.