Introduction
Influenza viruses are enveloped, single stranded RNA viruses in the family Orthomyxoviridae. The genome of influenza A viruses consists of eight unique segments of single-stranded RNA, which are of negative polarity (Webster et al., 1992). The hemagglutinin (HA) and neuraminidase (NA) are very important for the induction of an antibody response in the host, but they are also highly variable whereas the nucleoprotein (NP) and matrix (M) proteins are highly conserved between different influenza A viruses (Van Reeth, 2007). Nevertheless, influenza viruses are genetically unstable due to drift and shift antigenic mechanisms (Webster et al., 1992; Yassine et al., 2009). Influenza virions are pleomorphic, although their shape is generally roughly spherical with a diameter b150 nm. However, larger (100-400 nm) influenza virions are also generated as filamentous forms (Fujiyoshi et al., 1994).
Pigs play a crucial role in the interspecies transmission of influenza viruses (Horimoto and Kawaoka, 2001; Lipatov et al., 2004). They are susceptible to experimental infection by virtually any avian influenza strain (Kida et al., 1994), including viruses of the H5N1 subtype (Choi et al., 2005), and they are also susceptible to the strains circulating in humans, since porcine and human viruses are phylogenetically related, and they can easily cross the species barrier, as has happened with the new type A (H1N1) 2009 pandemic virus (Garten et al., 2009). In fact, phylogenetic and sero-archaeological studies suggest porcine involvement in the appearance of the strains that caused the human pandemics of the 20th century (Horimoto and Kawaoka, 2001).
The pathogenicity of influenza virus lies in its ability to elude host anti-viral immune responses. In mice, protection against influenza A virus requires that memory CD8+ T cells be reactivated by antigen presented by bone marrow derived dendritic cells (BMDCs) in the lymph nodes draining the site of infection (Castiglioni et al., 2008; Tamura and Kurata, 2004), while in pigs, primary influenza virus infection induced long-lived increase of lung CD8+ T cells and local lymphoproliferative responses (Charley et al., 2006). Activation of cell mediated immunity or cytotoxic T lymphocytes, in turn, depends on efficient delivery of signals by antigen presenting cells (APC). Dendritic cells (DCs), a heterogeneous population of haematopoietic cells, are the most potent APC in the body (Steinman, 2006). They play a versatile role in orchestrating immune responses against an array of invading pathogens, including influenza virus. The interactions between DCs and naïve and memory T cells determine both the magnitude and quality of the immune response. According to their functionality and phenotype, DCs can be classified as conventional DCs (cDCs) known as professional presenting cells or plasmacytoid DCs (pDCs), which naturally produce high levels of interferon type-I (Summerfield and McCullough, 2009). Indeed, it is well known that both cDCs and pDCs have important antigen-presenting functions and they complement each other by particular cross-talk pathways (Perez-Cabezas et al., 2011). Conventional DCs are amongst the first cells encountered by most viruses, simply due to their availability at every possible entry site of the body (Freer and Matteucci, 2009).
Extensive studies on DCs have been done in mice and humans and although the knowledge on swine immunology has been developing quickly in recent years, it is rather scarce. In mice, pDCs regulate the accumulation of T cells in the bronchoalveolar space during early influenza virus infection, but are dispensable for the control of this disease (Wolf et al., 2009). On other hand, the ability of human pDCs to engulf viral antigens from virus-containing cells without being infected, allows them to acquire and present viral antigens, avoiding virus-induced subversion of their functions (Lui et al., 2009). However, a recent study evaluated the ability of swine pDCs to sense different influenza viruses. In this study, authors found that pDC in one side may play a role in the cytokine storm observed during severe disease. On the other hand they could participate in early antiviral responses limiting virus replication (Michael et al., 2011). Also, previous results showed that influenza virus alters DC interaction with naïve T cells in vitro. Influenza virus infection increased the capacity of cDCs to stimulate T cell proliferation and infection is not required to observe enhanced T cell proliferation (Oh and Eichelberger, 1999). Functional disruption of conventional swine DCs has been described with another RNA virus, classical swine fever virus, as an important strategy for viral pathogens to evade host defences (Carrasco et al., 2004). Recently, Goldwich et al. (2011), studied theinteraction of herpes simplex virus type I (HSV-1) with human cDCs. Their results demonstrate that HSV-1 replicates in mature DCs but it can only be transferred to permissive cells in a cell-to-cell contact-dependent manner (Goldwich et al., 2011).
Lytic viruses such as influenza virus do not form a stable, longterm host-virus relationship within the infected host. Most of the studies of influenza viral particles formation have been performed on polarized epithelial cells (Nayak et al., 2009). In those studies, complete influenza virions were not present inside the infected cell and it could only be produced by budding from the plasma membrane. However, our knowledge about the roles of host factors in influenza infection is critically lacking, as well as the interaction of influenza viral particles with cells other than epithelial cells.
Given the pivotal role of DCs in triggering and directing the immune responses and the anatomical location of cDCs at the entry site of the body a set of experiments was designed to study the interaction between a circulating strain of porcine influenza virus (H3N2 SwIV) with swine cDCs in vitro. We found that influenzalike virions were located not only inside vesicles but also freely in the cytoplasm of influenza infected cDCs. Swine influenza virus interaction with cDCs did not induce viral budding nor detectable viral production in the supernatants. However, a limited increase in viral RNA was detected in cDCs by RT-qPCR. Surprisingly, H3N2 SwIV infected cDCs were able to infect susceptible cells when cell to cell contact was favoured. These new findings pave the way to elucidate the role of DCs in the course of influenza viral infection in its natural host.
Results
Ultrastructure and phenotype of poBMDCs
During the culture of porcine bone marrow haematopoietic cells with rpGM-CSF, cells grew in size, formed clusters and developed dendritic processes confirmed by electron microscopy at day eight (Figs. 1A, B, C). Porcine BMDCs in culture were semi-adherent cells, with some dendritic processes observed at day threewhich became noticeable as the culture progressed. At day eight of culture, the poBMDC phenotype was CD172a+, SLAI+, SLAII+, CD1+, CD4-, CD11R1-, CD14+, CD16+, CD40-, CD80/86+ and CD163low (Fig. 1D) which was consistent with previous reports (Carrasco et al., 2001; Kekarainen et al., 2008). At this time, the population of poBMDCs was rather homogenous with semi-mature cells and they constituted our starting culture for further experiments. Porcine BMDCs were also positive for α-2, 3 and α-2, 6 sialic acid (data not shown).
Infectious rate and viability of infected poBMDCs
Influenza viruses efficiently infect and replicate in vitro in epithelial cells for e.g. MDCK cells but no data was available concerning the viability of poBMDCs after H3N2 SwIV infection. Thus, levels of apoptotic versus necrotic cells were evaluated using different amounts of H3N2 SwIV in in vitro experiments to set up the experimental conditions for further analysis. Annexin V and propidium iodide staining were performed on poBMDCs 24 h after H3N2 SwIV infection. The overall mortality caused by H3N2 SwIV at 104 TCID50 per 106 cells was similar to that observed in mock treated cells, 16.5% and 12.5% respectively (Fig. 2A). When higher H3N2 SwIV doses were used, 105 TCID50 and 106 TCID50 per 106 cells, the percentage of dead cells increased to values around 30% of total cells (data not shown). Also, the percentage of apoptotic cells was similar to that of necrotic cells when 104 TCID50 was used (Fig. 2A).
In order to evaluate the percentage of poBMDCs actually infected by H3N2 SwIV when 104 TCID50 was used, intracellular NP staining of poBMDC was studied. Data in Fig. 2B shows that around 34% of poBMDCs were positive for influenza virus NP. Also, only cytoplasmic influenza virus NP staining was observed 24 h post-infection (hpi) in poBMDCs by immunofluorescence (Fig. 2C) whereas this staining was nuclear and cytoplasmic in the case of MDCKs at 24 hpi (data not shown). However, at 6 hpi both cell types exhibited nuclear staining for NP viral protein (data not shown). Subsequently, the amount of H3N2 SwIV used in the following experiments was 104 TCID50 for 106 poBMDCs in the cultures.
One of the hallmarks of DCs is their ability to up-regulate activation molecules after stimulation or infection. Consequently, it was investigated whether H3N2 SwIV- nfected poBMDCs exhibited any alteration in surface molecules such as SLA I, SLA II and CD80/86. A slight up-regulation with statistical tendency (p=0.06) in SLA I, II and CD80/86 expression was detected when values were compared with mock treated cells (Fig. 3). Values of mean fluorescence intensity were 22.7 for SLA I mock treated cells compared with 40.7 for H3N2 SwIV infected cells; for SLA II values were 40.6 compared to 64.6and for CD80/86 values were 8.5 compared to 11.8 respectively. Similar results of slight up-regulation, with statistical tendency (p=0.08) were observed in poBMDC stimulated for 24 h with poly:IC compared to mock treated cells (Fig. 3).
Interaction of H3N2 SwIV with poBMDCs versus MDCK
The replication cycle of influenza virus is composed of important steps such as fusion, endocytosis, replication, assembly and budding, which have been thoroughly investigated on permissive epithelial cells, like MDCK cells. Given the importance of DCs in triggering immune responses and once the conditions for H3N2 SwIV infection of poBMDCs were established, the H3N2 SwIV interaction with poBMDCs was analysed and compared with MDCK cells.
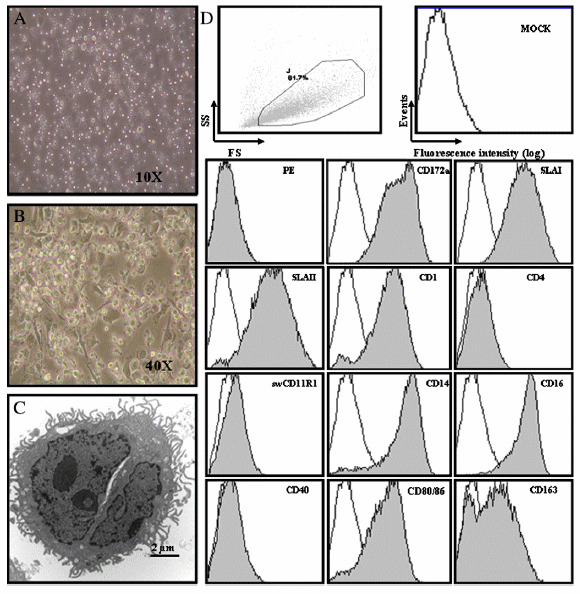
Fig. 1. Morphology and phenotype of poBMDCs. (A) poBMDCs seen by optical microscopy at day 3, 10× magnification. (B) Day 8 of generation 40× magnification and (C) electron microscopy at day 8 of generation. Bar=2 μm. (D) Gate strategy and phenotype of poBMDCs at day 8 of generation with rpGM-CSF. White histograms show isotype control stained cells and grey histograms represent the CD marker stained cells. Mock are poBMDCs only and isotype control are cells stained with the secondary antibody only. Representative results from nine independent experiments.
In the case of poBMDCs, structures resembling H3N2 SwIV virions were observed inside vesicles next to cellular membranes (Fig. 4A). Also, multiple vesicles were surprisingly observed in the cytoplasm, with 0.1 to 1 μm of diameter, containing several H3N2 SwIV like-particles with diameters of 80-100 nm. Most of them were surrounded by double membranes presenting H3N2 SwIV like-particles inside (Fig. 4B) which may reflect H3N2 SwIV particles entering the cell. Of note were several H3N2 SwIV like-particles observed free in the cytoplasm in close contact with Golgi complex. They may resemble virion structures budding from internal cistern of the Golgi complex membrane to the trans-Golgi network (Fig. 4C). Furthermore, large vesicles with several immature H3N2 SwIV like-particles, 70-80 nm in diameter, were observed in infected cells, some of them without capsids (Fig. 4D). In contrast with H3N2 SwIV-infected poBMDCs, in MDCK, H3N2 SwIV likeparticles were observed in the extracellular space next to the cellular membrane (Fig. 5A) and mature particles were observed in the extracellular space, next to and attached to the cellular membrane (Fig. 5B). In the cytoplasm of MDCKs, small and large vesicles were observed of less than 100 nm to 400 nm in diameter. These vesicles had simple or double membranes surrounded by immature (±80 nm) and mature (±100 nm) H3N2 SwIV like-particles in the cytosol (Fig. 5C). No H3N2 SwIV-like particles were detected freely in the cytoplasm of MDCK infected cells.
The structures inside poBMDCs resembled H3N2 SwIV but in order to assess whether they exhibited H3N2 SwIV proteins an immunogold labelling for H3N2 SwIV nucleoprotein coupled with 10 nm gold particles was performed on poBMDCs and MDCK infected cells. Those vesicles in the cytoplasm showed electron dense round and large structures consistent with H3N2 SwIV likeparticles of 80-100 nm in diameter in poBMDC cells (Figs. 4E, F). More importantly, these particles were heavily labelled. However, neither budding nor released virion particles were detected in poBMDCs. In MDCKs, electron dense structures with different size and irregular shape, surrounding nucleoli and chromatin were observed and labelled. Budding and released virions were labelled (Figs. 5D, E and F).
H3N2 SwIV infection of poBMDCs
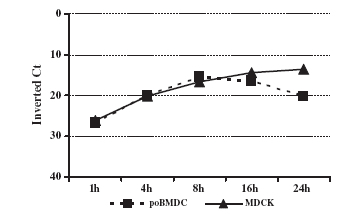
Generally, to evaluate influenza virus replication and generation of viral progeny, supernatant from infected cells is titrated on MDCKs. Supernatant from H3N2 SwIV infected-poBMDCs was assessed for viral progeny generation but no increase in viral titre was detected in poBMDCs compared with H3N2 SwIV-infected MDCKs at different time points. However, statistically significant differences between poBMDC and MDCK infected cells were observed at all time points with pb0.05 (Fig. 6). Therefore, the question was whether or not H3N2 SwIV was able to replicate in poBMDCs as compared with MDCK cells. Thus, 106 MDCK cells and 106 poBMDCs were infected in parallel using 104 TCID50, and viral RNA was evaluated by RT-qPCR at different time points. In MDCK cells, an increase in viral RNA was detected rising from 1 hpi to 24 h. In contrast, viral RNA in poBMDCs showed a limited increase between 1 and 8 hpi and later decayed with time. The increase in viral RNA was observed by comparing inverted Ct values. No significant differences were observed between poBMDC and MDCK infected cells (Fig. 7). The limit of detection in this assay was set at 30 Ct which corresponded to 103 TCID50 of H3N2 virus. Ct values for 107, 106, 105 and 104 TCID50 were 17, 19, 23 and 26 respectively.
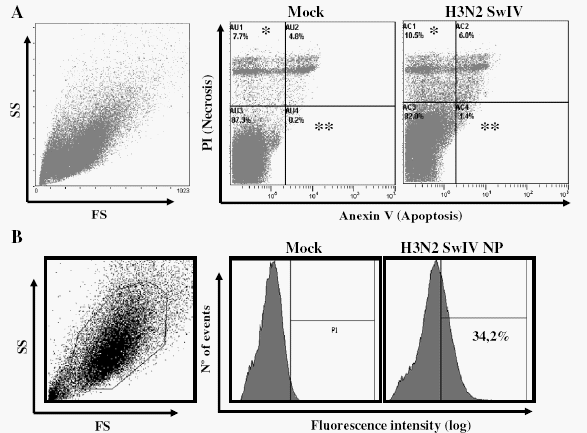
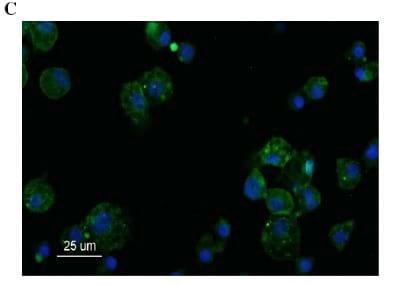
Fig. 2. Apoptosis/necrosis (A) and infectivity (nucleoprotein) (B) staining of poBMDCs after 24 h of infection with 0.01 MOI of SwIV H3N2. Mock or infected poBMDCs after 24 h were stained with (A) annexin V and/or propidium iodide, percentages in mock and infected cells were compared, *p=0.04, **p=0.01; n=5. The data was obtained without any gate strategy. (B) Anti-influenza NP staining in poBMDC by flow cytometry and (C) anti-influenza NP antibody (green) plus DAPI (blue) immunostaining of poBMDC. Bar=25 μm. FS (forward scatter) and SS (side scatter).
H3N2 SwIV infected poBMDCs are able to infect permissive cells by cell-to-cell contact
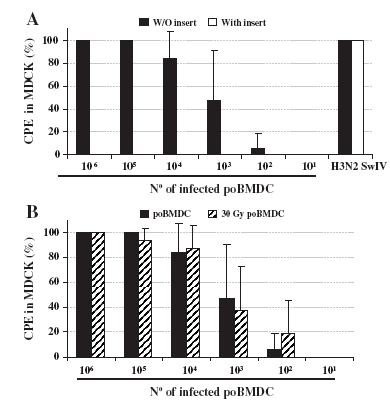
Once a limited increase of H3N2 SwIV RNA was detected in poBMDCs, the ability of H3N2 SwIV infected-poBMDCs to transmit the virus to other susceptible cells was evaluated. Thus, different amounts of H3N2 SwIV infected-poBMDCs were co-cultured with MDCK cells, in an infectious centre assay (ICA), in the absence or presence of a transwell (TA). When the TA was inserted and cell-to-cell contact was prevented, no cytopathic effect (CPE) was detected. In the absence of a TA insert, CPE on MDCK was observed in 100% of the wells when 106 to 105 infected poBMDCs were used. Subsequently, CPE levels fell when cell number decreased (Fig. 8A). One hundred microlitres of trypsin treated H3N2 5×105 TCID50 was also placed in the upper chamber of the transwell during the assay as a positive control. CPE was observed in 100% of the wells in this control (Fig. 8A). Moreover, high titres of virus were detected when supernatants from the 100% CPE positive wells in the ICA assay were titrated on MDCK cells. In these ICA positive wells, viral titres ranged from 105 to 106 TCID50/ml (data not shown). The question of whether influenza particles infecting susceptible cells originated from viral progeny in DCs or they were particles attached to the poBMDCs arose. In order to answer this question, poBMDCs and MDCK were irradiated with 30 Gy and 60 Gy respectively for 5 min before infection. Irradiation of MDCK before H3N2 SwIV infection was enough to prevent generation of infectious viral progeny in the supernatant (data not shown). After 24 h of infection, an ICA was performed. After 7 days in co-culture, either irradiated or non-irradiated poBMDC cells induced CPE on MDCK (Fig. 8B). The same result was obtained when irradiated MDCKs, infected with H3N2 SwIV, were tested (data not shown), which was not surprising considering the ability of SwIV viral particles to attach to the MDCK cell surface. Therefore, all the data suggested that viral particles attached to cDCs were able to infect susceptible cells only when cell-to-cell contact occurred.
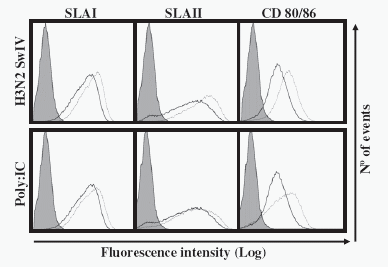
Fig. 3. Expression of SLA I, SLA II and CD80/86 24 h after H3N2 SwIV infection or 24 h after poly:IC stimulation. Infected/poly: IC stimulated-poBMDCs (dotted line) or uninfected/ unstimulated cells (continuous lines) were stained for SLA I, SLA II and CD80/86. Grey histograms represent the isotype control stained cells. Mean fluorescence of mock cells was compared with infected or poly:IC stimulated cells and a statistical tendency was found with p=0.06; n=3. The data was obtained with the same gate strategy shown in Fig. 1.
Discussion
The aim of this study was to evaluate the interaction between porcine DCs and a circulating SwIV and the possible role by the former in being carriers of porcine influenza virus. We used poBMDCs to strengthen our understanding of the interaction of SwIV with the host immune response. There is convincing evidence that cDCs play a critical role in vivo in immunity to acute infections (Castiglioni et al., 2008; Langlois and Legge, 2010; Montoya et al., 2005). Based on these studies, our working hypothesis was that SwIV infection of respiratory epithelial cells induces inflammation, resulting in recruitment of monocytes that differentiate into DCs and in activation of sentinel DCs. Then, resident DCs in the respiratory epithelial tissue would encounter SwIV during influenza virus infection. Indeed, DCs and macrophages reside beneath the epithelium of the respiratory organs, and these cells are potential targets for influenza viruses (Osterlund et al., 2010). Thus, DCs would be exposed to virus before leaving the inflamed tissue to stimulate the adaptive immune response in the regional lymph nodes. Porcine BMDCs are considered an experimental model for porcine cDCs, a group which includes monocyte derived DCs as well as resident DCs. Therefore, our studies were focused on understanding the nature of the interaction between H3N2 SwIV and cDCs in an in vitro system.
In a natural setting of influenza infection, humans and animals are usually infected at very low multiplicities of infection (MOI) (Nayak et al., 2004). Thus, our experimental system was set up using a rather low MOI (Fig. 2) with the intention of both, mimicking the natural setting of the infection and preserving cellular integrity during the experimental procedure.
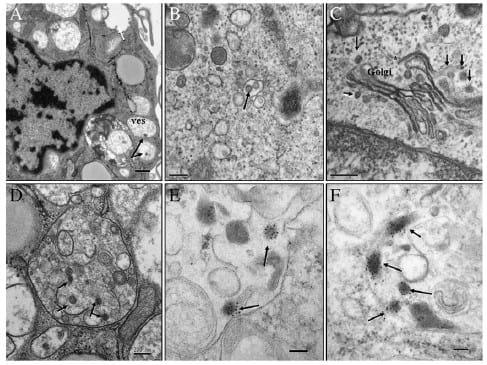
Fig. 4. H3N2-infected poBMDCs, 24 hpi. (A) Virions were located next to cellular membrane (black-arrows), inside vesicles (ves). A lot of vesicles were observed in the cytoplasm, with diameters of 0.1 to 1 μm (approx), containing several H3N2 SwIV like-particles of 80–100 nm in diameter (white-arrow). Bar=0.5 μm. (B) Most of the vesicles have double membranes surrounding H3N2 SwIV like-particles (arrow), bar=200 nm. (C) Next to the Golgi complex, several H3N2 SwIV like-particles were observed (arrow). Virion budding from internal cisternae of the Golgi complex membrane (*), to the trans-Golgi network. Bar=200 nm. (D) Large vesicle has several immature H3N2 SwIV like-particles with 70– 80 nm in diameter, and some of them without capsids (arrows). Bar=200 nm. (E, F) Immunogold labelling for H3N2 SwIV nucleoprotein coupled with 10 nm gold particle. Vesicles in the cytoplasm showing electron dense round and large structures, consistent with H3N2 SwIV like-articles with 80–100 nm in diameter, were heavily and specifically labelled (arrows). Bar=200 nm. Uranyl acetate and Reynolds lead citrate solution.
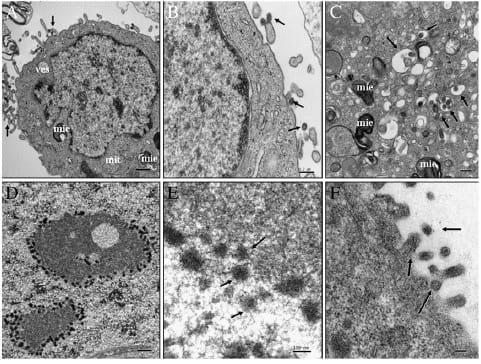
Fig. 5. H3N2-infected MDCK, 24 hpi. (A) These cells are characterized by large nuclei compared to their cytoplasm, where several large myelinoid figures (mie) and some vesicles were observed. H3N2 SwIV like-particles were detected in the extracellular space and next to cellular membrane (arrows). Bar=0.5 μm. (B) Close-up of figure A, mature H3N2 SwIV like-particles were observed in the extracellular space next to and attached to the cellular membrane (arrows). Bar=0.2 μm. (C) In the cytoplasm, small and large vesicles were observed of less than 100 nm to 400 nm diameter (approx). These vesicles contain SIV like-particles surrounded by simple or double membranes (±80 nm, arrows) and mature ones (±100 nm) in the cytosol (*). Bar=0.2 μm. (D, E) and (F) Immunogold labelling for H3N2 SwIV nucleoprotein coupled with 10 nm gold particle. Electron dense structures with irregular shape surrounding nucleoli and chromatin were observed and labelled (arrows). Budding release (F) (arrows) virions were labelled. Bar=0.5 μm, 100 nm and 100 nm respectively. Uranyl acetate and Reynolds lead citrate solution.
Interaction of DCs with virus has been proven to be extremely dependent on the type of virus and the DCs being studied. Our general knowledge on this interaction has been mainly obtained from humans and mice where there are numerous examples ranging from functional inhibition of DCs or weak response after viral infection (Osterlund et al., 2010) to the exploitation of DC functions for the dissemination of virus infection (Rowland-Jones, 1999). Other experimental systems have been used to study influenza infection and interaction with DCs in other natural hosts, like horses. In an in vitro equine monocyte-DC model, equine influenza virus successfully infected and initiated transcription of its viral genome in DCs, although only limited viral protein synthesis and progeny was achieved (Boliar and Chambers, 2010). In pigs, a study on porcine DCs has recently been published (Michael et al., 2011) but the authors focus on the ability of pDCs to respond to different influenza virus strains.
Therefore, our data on poBMDC interaction with a circulating H3N2 SwIV are, to our knowledge, the first one describing that porcine DCs support a limited increase in viral H3N2 SwIV RNA, and showing that influenza infected DCs are able to infect susceptible cells by cellto- cell contact.
In vitro, for different purposes including vaccine development, studies of influenza virus are mainly carried out in polarized cells (Youil et al., 2004). Thus, MDCK was used for many years as a model to study the replication cycle of influenza virus in vitro. A body of accumulating evidence indicates that complete influenza viral particles are not found inside the infected cell and the process of assembly, morphogenesis, budding and release of progeny virus particles takes place at the plasma membrane of the infected cells (Rossman and Lamb, 2011). These events are crucial for the production of infectious virions and pathogenesis of influenza virus. However, our data indicates that H3N2 SwIV like structures are detected freely in the cytoplasm of poBMDCs, surprisingly located near intracellular membranes (Golgi reticulum) (Figs. 4C, D, E and F). Similar experiments were conducted with other influenza viruses different from the H3N2 SwIV selected for this study to analyse whether this interaction was a particular feature of the H3N2 chosen. The results were identical to the ones shown previously with H3N2 SwIV in Fig. 4 (and data not shown). All these data suggest that SwIV interaction in porcine DCs might be different from the process taking place in polarized cells.
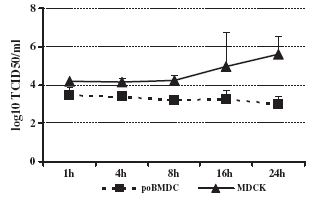
Fig. 6. H3N2 SwIV progeny in the supernatant of poBMDCs and MDCK infected cells. Supernatants from infected cells were collected at different time points after infection to titrate viral progeny on MDCK cells. Statistically significant differences between poBMDC and MDCK infected cells were observed at all time points with pb0.05. Bars represent the mean value plus one standard deviation; n=5.
Fig. 7. SwIV H3N2 RNA quantification in poBMDCs and MDCK infected cells. RT-qPCR was performed using viral RNA extracted from poBMDCs and MDCK infected cells at different times. No significant differences were observed between poBMDC and MDCK infected cells. Bars represent the mean value plus one standard deviation; n=3.
Fig. 8. Infectious centre and transwell assay. (A) Twenty-four hours post infection poBMDCs were co-cultured directly or indirectly with MDCK in 96 flat and 24 well plates respectively. As a positive control, 100 μl of trypsin treated H3N2 SwIV 5∗105 TCID50 was added. Bars represent the mean value plus one standard deviation; n=4. (B) Normal or irradiated poBMDCs with 30 Gy were infected for 24 h before the co-culture with MDCK in an ICA. CPE was evaluated at days 4 and 7 after ICA assay. No significant differences were observed between irradiated or not irradiated poBMDC. Bars represent the mean value plus one standard deviation; n=3.
Pioneering studies by Winkler and Cheville (1986) demonstrated that budding of pleomorphic virus particles from the alveolar epithelial cells and accumulation of viral protein within the nucleus and cytoplasm of epithelial cells were detected in lung sections from H1N1 SwIV infected pigs (Winkler and Cheville, 1986). Our studies in vitro also confirmthat budding of influenza particles is mainly seen in polarized epithelial cells (Fig. 5F) when compared with DCs in porcine cells.
In vivo, the involvement of other immune cells against influenza virus infection such as DCs has been less studied. During influenza virus infection, the role of DCs in priming an effective T cell response is crucial in generating adaptive responses. For influenza virus, RNA replication might be less effective on DCs than in other cell types. In fact, H3N2 SwIV infected-poBMDCs and MDCK (Figs. 6 and 7) exhibited marked differences in RNA viral production and replication, suggesting that this difference would probably account for such levels in vivo.
At early times post infection NP is localized predominantly to the nucleus, whereas at later times it is found in the cytoplasm, which reflects the trafficking of ribonucleoproteins during the virus life cycle. NP is primarily nuclear when expressed alone (Peter et al., 2007). At early times post infection (6 hpi) poBMDC andMDCK infected cells exhibited nuclear and cytoplasmic staining of influenza virus NP (data not shown). On the other hand, poBMDCs were positive for NP at 24 hpi but this staining was mainly detected in the cytoplasm of infected cells (Fig. 2C) whereas H3N2 SwIV-infected MDCKs exhibit NP staining in the nucleus as well as cytoplasm at this time point (data not shown). Also, infectious particles were not detected in the supernatant of poBMDC cultures (Fig. 6) and neither budding nor viral particle release was observed in poBMDCs by TEM (Fig. 4). All in all, this evidence would suggest that the limited increase in viral RNA is due to a default replication or an increase in influenza viral particles inside poBMDCs but no functional H3N2 SwIV replication was taking place in poBMDCs.
Additionally, infected poBMDCs were able to transmit H3N2 SwIV to susceptible cells in the ICA assays, indicating that virus transmission was efficient between both cells. However, when cell-to-cell contact was prevented between H3N2 infected poBMDCs and MDCK, no CPE was observed. H3N2 SwIV alone was able to induce CPE when an insert was used (Fig. 8A). The same behaviour was observed when H1N1 SwIV was used (data not show), indicating that the spread by cell-to-cell contact was not a particular signature of this H3N2 SwIV strain. Furthermore, when the cell replication machinery was inhibited by gamma-irradiation, H3N2 SwIV transmission from poBMDCs took place when susceptible cells were in close contact (Fig. 8B), indicating that H3N2 SwIV replication was not required for transmission of H3N2 SwIV from poBMDCs.
In humans, DCs serve as carriers of Human Immunodeficiency Virus (HIV) to the T cell area in the lymph nodes where T CD4+ cells are infected through CD4 and CCR5 receptors (Rowland-Jones, 1999). Based on the experimental data on HIV, DCs have been suggested to act as a "Trojan horses" for HIV in humans. In the porcine system, the fact that H3N2 SwIV infected poBMDCs were able to infect other susceptible cells when they are in close contact opens the possibility of speculation about the role of DCs carrying H3N2 SwIV infectious particles to other tissues than the respiratory organs. Dissemination of influenza virus in tissues outside the respiratory system has been described in experimentally infected mice (Sun et al., 2009) as well as in human patients suffering from severe influenza infection (Korteweg and Gu, 2008), however there is no evidence that SwIV could disseminate systemically in pigs. Alternatively, a plausible explanation for this finding would be that DC would carry the virus to secondary lymphoid organs such as lymph nodes where it could be detected by or possibly pass it on to B cells or other DCs for induction of specific immune responses (e.g. antibodies, CD8+ T cells...). This could be a beneficial effect to mount a specific immune response and thus contribute to viral clearance.
Finally, given the role of DCs in priming an effective and long lasting immune response, we report for the first time the interaction of porcine influenza viruses with poBMDCs. These data may help in understanding the role of DCs as important APC in the pathogenesis and epidemiology of influenza virus and the role of pigs as virus reservoirs. Taken together, our data shed new light on H3N2 SwIV interaction with poBMDCs. In contrast to earlier findings in other systems, an ability to infect susceptible cells by close contact was described. This opens new opportunities for the virus with respect to its dissemination inside the host and the interference with the host´s immune system at various sites besides the respiratory tract. Further studies have to be performed to elucidate the cause for this inefficient replication in porcine DCs and to define the consequences of influenza virus interaction with the immune system most potent APCs.
Materials and methods
Cells
Bone marrow haematopoietic cells were obtained from femurs of healthy Large white×Landrace pigs of eight weeks of age, negative for porcine reproductive and respiratory syndrome virus (PRRSV) and to type-2 porcine circovirus (PCV2) by RT-PCR as previously described (Olvera et al., 2004; Sibila et al., 2004). These animals were also negative by enzyme linked-immunosorbent assay (ELISA) for influenza virus and Actinobacillus (HIPRA, Amer, Spain), for mycoplasma (OXOID, Cambridge UK), for parvovirus, Adenovirus and Aujeszky´s disease virus (INGENASA, Madrid, Spain), and Salmonella (SVANOVA Biotech AB, Uppsala, Sweden). Bone marrow dendritic cells (BMDCs) were generated in an eight day protocol as previously described by Carrasco et al. (2001) with some modifications from Kekarainen et al. (2008). Briefly, bone marrow haematopoietic cells (BMHC) were resuspended in RPMI-1640 (Lonza, Walkesville, USA) culture medium containing 2 mM of L-glutamine (Invitrogen®, Barcelona, Spain), 100 U/ml of Polymyxin B (Sigma Aldrich Quimica S.A., Madrid, Spain), 10% of fetal calf serum (FCS) (Euroclone, Sziano,Italy) and 100 μg/ml of penicillin with 100 U/ml of streptomycin (Invitrogen®, Barcelona, Spain). This medium will be named RPMI-DC in this study. One hundred nanogrammes per millilitre (100 ng/ml) of recombinant porcine GM-CSF (R&D Systems, Spain) was added to the cells three times during the culture within 2 day intervals. Madin Darby Canine Kidney (MDCK) cells were maintained in a Dulbecco´s Modified Eagle Medium (DMEM) (Lonza, Walkesville, USA) containing 8 mM of L-glutamine and 200 μg/ml of penicillin with 200 U/ml of streptomycin and 5% of FCS. This medium will be referred as DMEM-MDCK in this study.
BMDC phenotype
Flow cytometry was performed using indirect labelling for CD172a, SLAI, SLAII, CD4, CD11R1, CD40, CD80/86 and CD163 and direct labelling for CD14 and CD16. Commercially available purified monoclonal antibodies (mAbs) anti-porcine CD14, CD16 and the fusion protein CD152 (CTLA4) for CD80/86 were used while the rest of the markers were detected by hybridoma supernatants. The secondary antibody was R-Phycoerythrin anti-mouse IgG (Jackson ImmunoResearch, Suffolk, UK). Briefly, 2.5∗105 cells/50 μl/well were labelled for 1 h at 4 °C for each CD marker, using 50 μl anti-CD172a (SWC3, BA1C11), 50 μl anti-SLA I (4B7/8), 50 μl anti-SLAII (1F12), 50 μl anti-CD1 (76-7-4), 50 μl anti-CD4 (76-12-4), and 50 μl anti- CD163 (2A10/11) and for anti-CD11R1 (MIL4, IgG1, Serotec), anti- CD14 (MIL2, IgG2b, Serotec, bioNova cientifica, Madrid, Spain), anti- CD16 (G7, IgG1, Serotec, bioNova cientifica, Madrid, Spain), purified anti-human CD40 (G28.4, IgG1κ, Biolegend, San Diego CA, USA), and CTLA4-mIg (Ancell, Minnesota, USA) the manufacturer´s instructions were followed. After 1 h of incubation at 4 °C, cells were washed with cold PBS with 2% FCS by centrifugation at 450 g, 4 °C for 10 min. Then, the secondary antibody R-Phycoerythrin diluted 1:200 was added when required. Cells were incubated for 1 h at 4 °C, and then they were washed as before and resuspended in PBS with 2% FCS. In order to determine if poBMDCs possess α-2,3-sialic acid or α-2,6 sialic acid linked to galactose, two lectins, MAA II (Maackia amurensis lectin II) and SNA (Sambucus nigra lectin) (Vector Laboratories, Peterborough, UK) with known capacities to bind α-2,3 or α-2,6 sialic acid respectively were chosen. Stained cells were acquired using a Coulter® EPICS XL-MCL cytometer and analysed by EXPO 32 ADC v.1.2 programme. A gate strategy was applied in 80% of living cells using the forward and side scatter (FS/SS) characteristic.
DC ultrastructure
At day eight, for conventional and immunogold labelling electron microscopy (EM) studies, poBMDCs were fixed with 2% (w/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde (EM grade, Merck, Darmstadt, Germany) in 0.1 M phosphate buffer (PB, Sigma-Aldrich, Steinheim, Germany), pH 7.4 and with 4% (w/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde in PB pH 7.4 respectively. For conventional EM procedures, cells were embedded in Eponate 12TM resin (Ted Pella, Inc, Redding, CA, USA). For immunogold labelling studies, after cryoprotection with sucrose (Sigma-Aldrich, Steinheim, Germany) solutions in PB at 4 °C, the pellet- ells were embedded in Lowicryl HM20 resin (Polysciences Inc., Warrington, USA). Briefly, the immunogold labelling was performed as follows: after blocking in 1% (w/v) bovine serum albumin (BSA; Sigma-Aldrich, Steinheim, Germany) in PBS (BSA/PBS), the grid samples were incubated with the monoclonal antibody to influenza A virus (Biodesign International, Saco, USA) at a dilution of 1:5 in 1% (w/v) BSA/PBS at 4 °C overnight in a humidified chamber. The secondary anti-mouse antibody was coupled to 10 nm-gold particles (British BioCell International, Cardiff, UK) in 1% (w/v) BSA/PBS for 40 min at room temperature.
Sections treated with PBS/BSA instead of primary antibody served as negative controls. All grid samples, from conventional and immunogold labelling studies, were contrasted with conventional uranyl acetate and Reynolds lead citrate solutions, and evaluated using a Jeol microscope 1400 (Jeol LTD, Tokyo, Japan) and photographed with a Gatan Ultrascan ES1000 CCD Camera 2048×2048 pixels as previously described by Rodríguez-Cariño et al. (2010) and Rodríguez-Cariño and Segales (2011).
Swine H3N2 influenza virus preparation and infection
Porcine A/Swine/Spain/SF32071/2007 (H3N2) SwIV strain was isolated from a natural outbreak on a conventional farm in Spain. Viral isolation was performed on embryonated specific pathogen free (SPF) eggs and subsequently multiplied on MDCK following the procedures of International Organization of Epizooties (OIE, 2008). Porcine BMDCs and MDCK were infected using a modified procedure, of previously described methodology by Rimmelzwaan et al. (1998). Briefly, 106 poBMDCs were infected with 104 TCID50 of previous porcine trypsin type IX (Sigma-Aldrich, St. Louis, USA) treated H3N2 107 TCID50/ml. Then, cells were incubated for 1 h at 37 °C 5% CO2 for virus adsorption. After this time, cells were thoroughly washed with PBS with 2% FCS and 400 μl of RPMI-DC was added.
For MDCK infection, DMEM medium supplemented with 8 mM of L-glutamine and 200 μg/ml of penicillin with 200 U/ml of streptomycin and 2 μg/ml of porcine trypsin type IX was added in the post infection DMEM (DMEM-PI). Mock and H3N2 SwIV infected cells were incubated for 1 h, 4 h, 8 h, 16 h and 24 h at 37 °C and 5% CO2. As control, poBMDCs were stimulated with 50 μg/ml of Polyinosinic Polycytidylic Acid Salt (Poly: IC) (Sigma Aldrich) for 24 h.
Percentage viability and intracellular staining for influenza nucleoprotein (NP)
Mortality 24 h after infection was determined by staining 2.5 ∗105 of mock or H3N2 SwIV infected cells with 5 μl of annexin V and after washing, adding 10 μl of propidium iodide (PI) (AB Serotec, Oxford, UK) following the manufacturer´s procedures.
At 24 hpi, intracellular staining for NP in poBMDCs was performed using 2.5 ∗105 cells fixed with 4% of paraformaldehyde (Electron Microscopy Science, Hatfield, PA, USA) for 30 min at 4 °C. After washing (centrifugation at 450 g for 5 min at 4 °C), cells were permeabilized with PBS with 0.2% v/v Tween 20 (Merck, Darmstadt, Germany) for 15 min at 37 °C. Then, cells were washed with PBS containing PBS+0.1% v/v Tween 20 and 100 μl of primary antibody of HB 65 (H16-L10-4R5-IgG2a) (ATCC® Manassas, USA) diluted 1:1000 in staining buffer (PBS with 0.1% w/v NaN3 and 1% w/v BSA) was added and incubated for 1 h at 4 °C. After washing, cells were incubated with 1:200 diluted fluorescein (FITC) conjugated affinity pure F(ab′)2 fragment goat anti-mouse IgG (Jackson ImmunoResearch, Suffolk, UK), for 1 h at 4 °C. Then, after washing, cells were resuspended in staining buffer and analysed by FACSaria (Becton Dickinson). After 24 h of infection or poly:IC stimulation, poBMDCs were harvested and stained as described elsewhere for SLA I, SLA II and CD80/86 using monoclonal antibodies or immunoglobulin fusion protein.
Immunofluorescence of influenza virus NP
Presence of NP viral protein in poBMDCs wasvisualized by indirect immunofluorescence in infected cells. At 24 hpi, mock or infected poBMDCs were placed on a circular glass cover slip (VWR International, Spain) and left to adhere for 1 h at 37 °C using 40 μl of fibronectin from human plasma at 20 μg/ml (Sigma Aldrich). After that, cells were fixed with ethanol (Panreac, Spain) for 10 min at 4 °C, dehydrated with acetone and permeabilized with 0.1% of Triton X-100 for 15 min at 37 °C. Then, cells were washed with PBS 2% FCS and 100 μl of primary antibody HB 65 (H16-L10-4R5-IgG2a) (ATCC® Manassas, USA) diluted 1:500 was added. Cover slips were incubated at 4 °C for 1 h and after two rounds of washes, 100 μl of CyTM 2-goat anti-mouse IgG (Jackson Immunoresearch, Suffolk, UK) diluted 1:200 was added for a further 1 h of incubation at 4 °C. Finally and after several washes, nuclei were counterstained with DAPI. Cover slips were dried and mounted using 1 drop of Fluoprep (BioMérieux, France). To detect autofluorescence, mock or infected-poBMDCs, were stained as controls with the primary and/or secondary antibody. Treated cells were viewed on a Nikon eclipse 90i epifluorescence microscope equipped with a DXM 1200F camera (Nikon Corporation, Japan). Pictures were merged using Adobe®Photoshop®CS version 8 (Adobe System Incorporated, USA).
H3N2 replication in BMDCs and MDCK cells
Virus replication in infected cells was assessed by titration of supernatants on MDCK cells with the aid of trypsin in the post-infection media. Virus titre was calculated by the Reed and Muench method (Reed and Muench, 1938). Viral threshold cycle (Ct) values in MDCK or BMDC cells were assessed following a TaqMan one-stepquantitative RT-PCR (RT-qPCR) in Fast7500 equipment (Applied Biosystems, Foster City, CA). RT-qPCR was performed using 60 μl of eluted RNA extracted from mock or infected cells using TRIZOL® reagent (Invitrogen®, San Diego, USA). The primers and probe and the amplification conditions used to perform the RT-qPCR were previously described by Busquets et al. (2010). The amplification profile was as follows: reverse transcription at 48 °C for 30 min; initial denaturation reaction at 95 °C for 15 min and 40 PCR-cycles of 95 °C for 15 s and 60 °C for 1 min. Serial 10-fold dilutions of H3N2 RNA, obtained from H3N2 infected MDCK of known concentration, were made and a standard curve generated. The limit of detection was 103 TCID50/ml corresponding to the Ct 30.
Infectious centre assay (ICA) and transwell assay (TA)
At 24 hpi, mock or H3N2 SwIV infected-DCs ranging from 106 to 10 cells were co-cultured with MDCK in the presence of DMEM-PI in 96 (Nunc® Kamstrupvej, Denmark) or in BD Falcon cell culture 24-well plate with or without inserts with 0.4 μm pores (Becton Dickinson) respectively. Then as positive control, 100 μl of trypsin treated H3N2 5∗105 TCID50 was added in the TA assay. In addition, poBMDCs and MDCK were irradiated for 5 min with 30 Gy and 60 Gy respectively in an IBL 437C type H irradiator (CIS, Biointernational, Nice, France) before infection. After irradiation, cells were washed and infected with H3N2 SwIV for 1 h at 37 °C (for virus adsorption). Then, cells were washed and incubated for 24 h. After 24 hpi, cells were washed, counted and co-cultured with MDCK for 7 days in an ICA. At days four and eight after ICA or TA assays cytopathic effect (CPE) in MDCK was evaluated. Each condition in the co-culture had 8 replicas in the 96-well plate. Then, CPE was quantified as being positive when 70% to 100% of monolayer disruption was observed in the wells from the 96-well plate with MDCK. When less than 70% percentage was observed, the well was considered negative.
Statistical analysis
All statistical analyses were carried out using the SAS system V.9.1.3 (SAS Institute Inc, Cary, NC, USA). The significance level (α) was set at 0.05 with statistical tendencies reported when p<0.10. A non-parametric test (Mann-Whitney) was used to compare any variable response between experimental groups. In the particular case of the cytopathic effect in MDCK cells, an ANOVA test was carried out using a number of infected poBMDCs and cell irradiation as independent variables.
Acknowledgments
This work was partly funded by the following projects: CSD 2006-00007, AGL2006-13809-C03-01, AGL2009-12945-C02-01 and AGL2010-22200-C02-01 by the Spanish Government. PhD studies of Mrs. Tufária Mussá and Elisa Crisci are supported by a doctoral grant from the AECID and from the Spanish Ministry of Science and Innovation respectively. Authors also thank Alejandro Sanchez Chardi from Electronic Microscopy Unit at Universitat Autònoma de Barcelona for his kind support and the DC.CAT group (the Catalan group for DCs studies) and Dr. Llilianne Ganges for suggestions and critically reviewing the manuscript.
References
Boliar, S., Chambers, T.M., 2010. A new strategy of immune evasion by influenza A virus: inhibition of monocyte differentiation into dendritic cells. Vet. Immunol. Immunopathol. 136 (3-4), 201-210.
Busquets, N., Segales, J., Cordoba, L., Mussa, T., Crisci, E., Martin-Valls, G.E., Simon-Grife, M., Perez-Simo, M., Perez-Maillo, M., Nunez, J.I., Abad, F.X., Fraile, L., Pina, S., Majo, N., Bensaid, A., Domingo, M., Montoya, M., 2010. Experimental infection with H1N1 European swine influenza virus protects pigs from an infection with the 2009 pandemic H1N1 human influenza virus. Vet. Res. 41 (5), 74.
Carrasco, C.P., Rigden, R.C., Schaffner, R., Gerber, H., Neuhaus, V., Inumaru, S., Takamatsu, H., Bertoni, G., McCullough, K.C., Summerfield, A., 2001. Porcine dendritic cells generated in vitro: morphological, phenotypic and functional properties. Immunology 104 (2), 175-184.
Carrasco, C.P., Rigden, R.C., Vincent, I.E., Balmelli, C., Ceppi, M., Bauhofer, O., Tache, V., Hjertner, B., McNeilly, F., van Gennip, H.G., McCullough, K.C., Summerfield, A., 2004. Interaction of classical swine fever virus with dendritic cells. J. Gen. Virol. 85 (Pt 6), 1633-1641.
Castiglioni, P., Hall de, S., Jacovetty, E.L., Ingulli, E., Zanetti, M., 2008. Protection against influenza A virus by memory CD8 T cells requires reactivation by bone marrowderived dendritic cells. J. Immunol. 180 (7), 4956-4964.
Charley, B., Riffault, S., Van Reeth, K., 2006. Porcine innate and adaptative immune responses to influenza and coronavirus infections. Ann. N. Y. Acad. Sci. 1081, 130- 136.
Choi, Y.K., Nguyen, T.D., Ozaki, H., Webby, R.J., Puthavathana, P., Buranathal, C., Chaisingh, A., Auewarakul, P., Hanh, N.T., Ma, S.K., Hui, P.Y., Guan, Y., Peiris, J.S., Webster, R.G., 2005. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J. Virol. 79 (16), 10821-10825.
Freer, G., Matteucci, D., 2009. Influence of dendritic cells on viral pathogenicity. PLoS Pathog. 5 (7), e1000384.
Fujiyoshi, Y., Nahoaki, K., Sakata, P., Kazumi, Sato, S.B., 1994. Fine structure of influenza A virus observed by electron cryo-microscopy. EMBO J. 13 (2), 318-326.
Garten, R.J., Davis, C.T., Russell, C.A., Shu, B., Lindstrom, S., Balish, A., Sessions, W.M., Xu, X., Skepner, E., Deyde, V., Okomo-Adhiambo, M., Gubareva, L., Barnes, J., Smith, C.B., Emery, S.L., Hillman, M.J., Rivailler, P., Smagala, J., de Graaf, M., Burke, D.F., Fouchier, R.A., Pappas, C., Alpuche-Aranda, C.M., Lopez-Gatell, H., Olivera, H., Lopez, I., Myers, C.A., Faix, D., Blair, P.J., Yu, C., Keene, K.M., Dotson Jr., P.D., Boxrud, D., Sambol, A.R., Abid, S.H., St George, K., Bannerman, T., Moore, A.L., Stringer, D.J., Blevins, P., Demmler-Harrison, G.J., Ginsberg, M., Kriner, P., Waterman, S., Smole, S., Guevara, H.F., Belongia, E.A., Clark, P.A., Beatrice, S.T., Donis, R., Katz, J., Finelli, L., Bridges, C.B., Shaw, M., Jernigan, D.B., Uyeki, T.M., Smith, D.J., Klimov, A.I., Cox, N.J., 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325 (5937), 197-201.
Goldwich, A., Prechtel, A.T., Muhl-Zurbes, P., Pangratz, N.M., Stossel, H., Romani, N., Steinkasserer, A., Kummer, M., 2011. Herpes simplex virus type I (HSV-1) replicates in mature dendritic cells but can only be transferred in a cell-cell contact-dependent manner. J. Leukoc. Biol. 89 (6), 973-979.
Horimoto, T., Kawaoka, Y., 2001. Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14 (1), 129-149.
Kekarainen, T., Montoya, M., Dominguez, J., Mateu, E., Segales, J., 2008. Porcine circovirus type 2 (PCV2) viral components immunomodulate recall antigen responses. Vet. Immunol. Immunopathol. 124 (1-2), 41-49.
Kida, H., Ito, T., Yasuda, J., Shimizu, Y., Itakura, C., Shortridge, K.F., Kawaoka, Y., Webster, R.G., 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75 (Pt 9), 2183-2188.
Korteweg, C., Gu, J., 2008. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 172 (5), 1155-1170.
Langlois, R.A., Legge, K.L., 2010. Plasmacytoid dendritic cells enhance mortality during lethal influenza infections by eliminating virus-specific CD8 T cells. J. Immunol. 184 (8), 4440-4446.
Lipatov, A.S., Govorkova, E.A., Webby, R.J., Ozaki, H., Peiris, M., Guan, Y., Poon, L., Webster, R.G., 2004. Influenza: emergence and control. J. Virol. 78 (17), 8951-8959.
Lui, G., Manches, O., Angel, J., Molens, J.P., Chaperot, L., Plumas, J., 2009. Plasmacytoid dendritic cells capture and cross-present viral antigens from influenza-virus exposed cells. PLoS One 4 (9), e7111.
Michael, B., Manuela, O.-M., Matthias, L., Kenneth, C.M., Mikhail,, M.a.A.S., 2011. Efficient sensing of avian influenza viruses by porcine plasmacytoid dendritic cells. Viruses 3, 312-330.
Montoya, M., Edwards, M.J., Reid, D.M., Borrow, P., 2005. Rapid activation of spleen dendritic cell subsets following lymphocytic choriomeningitis virus infection of mice: analysis of the involvement of type 1 IFN. J. Immunol. 174 (4), 1851-1861.
Nayak, D.P., Hui, E.K., Barman, S., 2004. Assembly and budding of influenza virus. Virus Res. 106 (2), 147-165.
Nayak, D.P., Balogun, R.A., Yamada, H., Zhou, Z.H., Barman, S., 2009. Influenza virus morphogenesis and budding. Virus Res. 143 (2), 147-161.
Oh, S., Eichelberger, M.C., 1999. Influenza virus neuraminidase alters allogeneic T cell proliferation. Virology 264 (2), 427-435.
OIE, 2008. Manual of diagnostic tests and vaccines for terrestrial animals. http://www. oie.int/eng/normes/mmanual/2008/pdf/2.08.08_SWINE_INFLUENZA.pdf,2.8. Olvera, A., Sibila, M., Calsamiglia, M., Segales, J., Domingo, M., 2004. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J. Virol. Meth. 117 (1), 75-80.
Osterlund, P., Pirhonen, J., Ikonen, N., Ronkko, E., Strengell, M., Makela, S.M., Broman, M., Hamming, O.J., Hartmann, R., Ziegler, T., Julkunen, I., 2010. Pandemic H1N1 2009 influenza A virus induces weak cytokine responses in human macrophages and dendritic cells and is highly sensitive to the antiviral actions of interferons. J. Virol. 84 (3), 1414-1422.
Perez-Cabezas, B., Naranjo-Gomez, M., Bastos-Amador, P., Requena-Fernandez, G., Pujol- Borrell, R., Borras, F.E., 2011. Ligation of Notch Receptors in Human Conventional and Plasmacytoid Dendritic Cells Differentially Regulates Cytokine and Chemokine Secretion and Modulates Th Cell Polarization. J. Immunol. 186 (12), 7006-7015.
Peter, P., Shaw, Megan L., Howley, Peter M., 2007. Orthomyxoviridae: the viruses and their replication, In: Knipe, David, M. (Eds.), Fields Virology, 5th Edition. Lippincott Williams & Wilkins, Philadelphia, pp. 1648-1660.
Reed, L.J., Muench, H., 1938. A simple method of estimating fifty per cent endpoint. Am. J. Hyg. 27, 493-497.
Rimmelzwaan, G.F., Baars, M., Claas, E.C., Osterhaus, A.D., 1998. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods 74 (1), 57-66.
Rodriguez-Carino, C., Sanchez-Chardi, A., Segales, J., 2010. Subcellular immunolocalization of porcine circovirus type 2 (PCV2) in lymph nodes from pigs withpostweaning multisystemic wasting syndrome (PMWS). J. Comp. Pathol. 14 (4), 291-299.
Rodriguez-Carino, C., Duffy, C., Sanchez-Chardi, A., McNeilly, F., Allan, G.M., Segales, J., 2011. Porcine circovirus type 2 morphogenesis in a clone derived from the l35 lymphoblastoid cell line. J. Comp. Pathol. 144 (2-3), 91-102.
Rossman, J.S., Lamb, R.A., 2011. Influenza virus assembly and budding. Virology 411 (2), 229-236.
Rowland-Jones, S.L., 1999. HIV: the deadly passenger in dendritic cells. Curr. Biol. 9 (7), R248-R250.
Sibila, M., Calsamiglia, M., Segales, J., Blanchard, P., Badiella, L., Le Dimna, M., Jestin, A., Domingo, M., 2004. Use of a polymerase chain reaction assay and an ELISA to monitor porcine circovirus type 2 infection in pigs from farms with and without postweaning multisystemic wasting syndrome. Am. J. Vet. Res. 65 (1), 88-92.
Steinman, R.M., 2006. Linking innate to adaptive immunity through dendritic cells. Novartis Found. Symp. 279, 101-109 discussion 109-13, 216-9.
Summerfield, A., McCullough, K.C., 2009. The porcine dendritic cell family. Dev. Comp. Immunol. 33 (3), 299-309.
Sun, R., Luo, J., Gao, Y., He, H., 2009. Different infection routes of avian influenza A (H5N1) virus in mice. Integr Zool. 4 (4), 402-408.
Tamura, S., Kurata, T., 2004. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn. J. Infect. Dis. 57 (6), 236-247.
Van Reeth, K., 2007. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet. Res. 38 (2), 243-260.
Webster, R.G., Bean, W.J., Gorman, O.T., Chambers, T.M., Kawaoka, Y., 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56 (1), 152-179.
Winkler, G.C., Cheville, N.F., 1986. Ultrastructural morphometric investigation of early lesions in the pulmonary alveolar region of pigs during experimental swine influenza infection. Am. J. Pathol. 122 (3), 541-552.
Wolf, A.I., Buehler, D., Hensley, S.E., Cavanagh, L.L., Wherry, E.J., Kastner, P., Chan, S., Weninger, W., 2009. Plasmacytoid dendritic cells are dispensable during primary influenza virus infection. J. Immunol. 182 (2), 871-879.
Yassine, H.M., Khatri, M., Zhang, Y.J., Lee, C.W., Byrum, B.A., O´Quin, J., Smith, K.A., Saif, Y.M., 2009. Characterization of triple reassortant H1N1 influenza A viruses from swine in Ohio. Vet. Microbiol. Youil, R., Su, Q., Toner, T.J., Szymkowiak, C., Kwan, W.S., Rubin, B., Petrukhin, L., Kiseleva, I., Shaw, A.R., DiStefano, D., 2004. Comparative study of influenza virus replication in Vero and MDCK cell lines. J Virol Methods 120 (1), 23-31.
This article was originally published by Elsevier, Virology 420 (2011) 125–134 in October, 2011. Engormix.com thanks for this contribution.