Moving horse genomics across disciplinary lines
Published: July 16, 2007
By: ERNEST BAILEY (Courtesy of Alltech Inc.)
One of the great ironies for agriculture is the popular recognition horse breeders enjoy as genetics experts while academics in agriculture consider horse breeding an obtuse art. Indeed, even horse breeders prefer to regard breeding as an art and every horse a canvas.
It is an apt analogy, because one way to be regarded a great horse breeder is one masterpiece, a Kentucky Derby winner. In practice, of course, those who achieve that goal have been long-term practitioners of the art and are very deserving.
Horses were once a more important part of agriculture and highly selected to provide power, draft and transportation. With the advent of the modern combustion engine and other energy sources, the horse has become more important for recreation and unimportant as a source of power for industry and agriculture.
Although the horse does not contribute to the ‘food and fiber’ mission of agriculture, the horse industry still has a large impact on the economy. The results of a recent study by the American Horse Council (AHC, 2005) can be seen in Table 1.
Table 1. Economic impact of the horse industry in the United States.
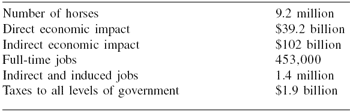
The horse industry provides jobs and money to the American economy and has a comparable impact worldwide. Management practices and research that make the horse business more effective benefit our economy. Horse owners value horses that are athletic, healthy, intelligent and responsive.
Genetic selection and effective management are valuable tools; and genomics research on horses can enhance both selection and management. Genomics is transforming the way that we do research in agriculture. (Here, genomics is being used in a broad sense to include genomics, transcriptomics, proteomics, metabalomics, etc.)
Management practices and the foodstuff we provide our livestock influence the array of genes that are expressed throughout the tissues of the horse, even without consideration of hereditary potential.
Why agriculturalists support animal genomics
During the last 100 years, geneticists have accomplished amazing feats in food animal agriculture through genetic selection. Within the last 50 years alone, dairy cattle have doubled milk yields because of selection. Using the record-keeping model used by dairymen, meat producers made significant advances with beef, pork and poultry during the same period. Of course, these accomplishments were made without regard to DNA.
However, agricultural scientists are pragmatic if nothing else. In the late 1980s, a quantitative geneticist named Morris Soller projected the gains that one might realistically anticipate for the next 25 years using traditional quantitative genetics breeding methods for milk production.
Next, he projected the gain that one might obtain under the best circumstances following molecular biologists’ discovery of a gene with a major effect on production and consequent selection over the same 25 year period.
The results: quantitative genetics would win, no question. Still, this eminent quantitative geneticist advocated pursuing molecular genetics studies of production traits in livestock. Why? Because he also projected that one day the gains made using quantitative genetics would peak and that future advances would require an understanding of the molecular mechanisms underlying hereditary traits.
A strategy and rationale for applying markerassisted selection was described in a subsequent publication (Weller et al., 1990). Future advances in production would come through a comprehensive understanding of molecular genetics, a field now referred to as genomics.
Thumbnail history of the human genome sequencing projects
Genomics came to the fore of popular knowledge through the Human Genome Project (HGP). In October 1990 an international plan was established to fully sequence the human genome over a 15 year period. It was a bold plan, because at the time the technology did not exist to sequence and assemble a large complex genome.
Indeed, much of the funding and work was directed at developing the technologies. In May 1998, a private company (Celera) was formed to compete with the international effort and complete the human genome ahead of the HGP schedule. Theirs was a bold plan because it relied on computing power and computer programs that did not exist at the time.
Ultimately the two programs collaborated and announced a tie finish in the spring of 2003, coincidental with the 50th anniversary of Watson and Crick’s report on the structure of DNA. The applications of the information appear almost daily in our newspapers and have revolutionized medicine. The newly invented technologies also had a large impact in other arenas including agriculture and environmental sciences. We entered the ‘biology century’ (http://www.ornl.gov/sci/techresources/Human_Genome/ home.shtml).
History of animal genome projects
The applications in animal agriculture began in the early 1990s. In 1992, the United States Department of Agriculture (USDA) established the National Research Sponsored Project (NRSP) No. 8 in which monies were taken directly out of state appropriations for support of agricultural colleges to develop genomic studies for agriculturally important animals.
Scientists from land grant institutions in the United States along with scientists from industry and from around the world collaborated to create gene maps for livestock. The idea was to identify genes having a major effect on production traits or having a major impact on health and welfare of the animals.
From 1992 to 1997, NRSP8 included cattle, sheep, pigs and chickens. By 1997, scientists organized workshops on horse genomics and joined NRSP8 while aquaculture scientists joined NRSP8 in 2003. NRSP8 provides funds to coordinate collaborative research and meetings are held annually to discuss research advances and make plans for future collaborations.
NRSP8 has been one of the most successful collaborative efforts in agricultural research; effective gene maps have been created for all species and molecular determinants of health and production identified and diagnostic tests developed.
SEQUENCING THE GENOMES OF LIVESTOCK SPECIES
With completion of the human genome project, resources became available to sequence the genomes of other animal species. So far, among agriculturally important species we have completed genome sequences for chickens and cattle. Sequencing is underway for sheep and pigs. The initial projection for the human genome project was $3 billion.
However, that estimate included invention of technologies. The Celera project to sequence the human genome in 1998 was projected at a mere $300 million. Likewise, they had to invent technologies. The cost for sequencing the chicken genome was $60 million.
Cattle genome sequencing a few years later cost $52 million. Today, a low level sequencing is being done for pigs and sheep that identify only 85-90% of the genes at a cost of $10 million. Costs are going down and the assembly of the genomes is becoming easier as more species are sequenced and genome order assembled.
Genome organization is very similar among vertebrate species. Indeed, one of the major programs for sequencing the genomes of mammalian species is that of the National Human Genome Research Institute (NHGRI) (http://www.genome.gov/12511814). NHGRI has a program to sequence the genomes of diverse species in connection with annotation of the human genome.
In that program they are conducting low level DNA sequencing for diverse species including mice, rats, dogs, elephants, tree shrews, marsupials, tenrecs, cats, armadillos, rabbits, hedgehogs and guinea pigs.
As of February 2006, the horse has been added to that list. Comparison of the genome sequences of these different species helps to identify conserved regulatory regions in the DNA. These regulatory regions could not be found otherwise and the relevance of the large stretches of repetitive DNA sequences remains mysterious.
What we know about mammalian genomes
The genomics revolution revealed that all mammals possess approximately 20,000 pairs of genes. Within a cell, 10,000 of those genes will be expressed. The particular combination of 10,000 genes is characteristic of the tissue and its metabolic state. Genes may be up-regulated or down-regulated.
Investigation of gene expression is an area of genomics called functional genomics or transcriptomics. Gene expression is commonly assayed by Northern blots or by use of microarray assays allowing simultaneous evaluation of all possible genes. At the same time, genes have an intron-exon structure that allows expression of several structural versions.
Curiously, these 20,000 genes represent only 3% of the DNA in a cell. The remainder (97%) has been called ‘junk DNA’, but more because of our ignorance than a qualitative evaluation of the DNA.
Indeed, comparisons of DNA among diverse species reveals that vast stretches of DNA have been conserved over 100 million years of evolution (poultry to humans) and probably represent important regulatory DNA elements. This DNA is less conserved than genes but more conserved than random expectations.
Another surprising discovery was the conservation of gene order among vertebrates. Consequently, we can use the gene order in one species to predict the order in another. When we map a trait to a particular chromosome region and know its homology to the human genome organization, we can use the human genome sequence to predict candidate genes in livestock. Identifying candidate genes is really the first step in identifying gene mutations.
The low number of genes and their conservation among species is useful knowledge. The consequence of this observation for agriculture is that when a gene is found to cause a disease in humans, a similar disease in livestock is likely to be the product of a homologous gene.
We can use the information from human genomics to predict which genes are going to have an effect in livestock. For horses, this approach led to the discovery of the cause of hyperkalemic periodic paralysis (HYPP) in Quarter horses (Rudolph et al., 1992), severe combined immunodeficiency disease (SCID) in Arabian horses (Shin et al., 1997) and overo lethal white foal syndrome (OLWFS) in Paint horses (Metallinos et al., 1998; Santschi et al., 1998; Yang et al., 1998).
Genomics and horses
In 1995 a workshop was formed among scientists from among 25 laboratories working on problems related to genetics in horses (http://www.uky.edu/AG/horsemap). No one laboratory had all the resources necessary to complete a gene map for the horse; and this was the tool that each of the scientists needed to pursue their research into musculoskeletal diseases, infectious diseases, hereditary diseases, behavior, performance and even coat color patterns.
Indeed, by the 1990s it was clear that biological research on an organism would require a comprehensive gene map (By 2005 that requirement became a whole genome sequence!).
Over the next decade an agreement was reached on nomenclature for chromosomes (ISCNH, 1997), several linkage maps were reported (Lindgren et al., 1998; Guerin et al. 1999; Swinburne et al., 2000; Penedo et al., 2005; Swinburne et al., 2006), a radiation hybrid map was published (Chowdhary et al., 2003), and numerous other independent research projects filled in gaps, corrected technical errors and increased the efficacy of the horse gene map for identifying hereditary traits.
By 2006, 1000- 1500 genetic markers have been linkage mapped; more than 1200 genes have been mapped using cytogenetics and radiation hybrid techniques and we anticipate publication of a new radiation hybrid map within the next year that includes more than 4000 genes and genetic markers (Chowdhary, personal communication, January 2006).
As the map developed, it was used to identify genes and chromosome regions encoding hereditary traits in horses. The initial focus of the research efforts has been on simple Mendelian traits. The list of traits that have been mapped is shown in Table 2.
Table 2.Hereditary traits identified or mapped for the horse.
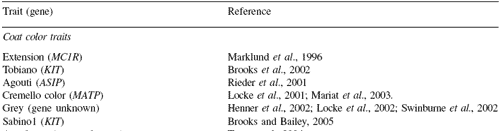

Discovering the genes for the traits listed in Table 2 represents the horse genetics equivalent of ‘harvesting the low-hanging fruit’. Each of the traits was clearly hereditary in nature.
Besides creating diagnostic tests for the traits, the most important aspects of these studies were to demonstrate that using horse genomics information could be effective to better characterize hereditary traits.
However, we need more powerful genomics tools to understand and resolve management questions for more complex traits in horses. For example, we anticipate that some musculoskeletal diseases of horses will involve multiple genetic systems, nutritional factors and management. Understanding such traits will require expertise from several disciplines. Indeed, most of the traits valued by horse breeders and horse owners are complex traits that combine selection, nutrition, training and other aspects of management.
Horses are valued for athleticism of event horses, strength of draft horses, strength and size of draft ponies, temperament, gait and, of course, racing performance of Thoroughbred horses.
The horse genome will be sequenced and assembled during 2006 by the NHGRI. The additional power from this information will have a dramatic impact on genomics tools. We can anticipate the following developments in the arena of genomics.
INVESTIGATIONS OF GENE EXPRESSION (TRANSCRIPTOMICS)
Currently, we have equine cDNA microarrays in development that allow us to assess 8,000 of the 20,000 horse genes.
The genome sequence will make it possible to create oligonucleotide arrays that assay expression of all 20,000 genes based on having the entire genome sequence. This means that nutritional studies can address the effect of nutrients on biochemical pathways or single genes.
As a horse is trained, as a horse recovers from disease and as a horse is inherently constituted, a unique set of genes will be expressed in its cells. Assaying the particular set of genes being expressed may provide valuable information for making choices about management, training and even selection.
The 20,000 element microarray will be a complex research tool, but as we use it to better understand nutrition and disease processes, this research tool will give way to many diagnostic tools.
GENETIC MARKER STUDIES
Currently we have a gene map with 1000 to 1500 genetic markers. Genetic studies rely on the use of several hundred mapped microsatellite markers that are segregated into families. The whole genome sequence will make it possible for us to assay tens of thousands of genetic markers called single nucleotide polymorphisms (SNPs).
Based on the large number of markers and with knowledge of the breed history, we can assay affected and unaffected individuals for a pattern of genetic variation at the population level.
This will be a particular boon because horse breeders may be reluctant to continue breeding horses suspected of having hereditary predispositions to disease. Families segregating for some traits are hard to find so we need to develop a strategy independent of family structure.
It is an apt analogy, because one way to be regarded a great horse breeder is one masterpiece, a Kentucky Derby winner. In practice, of course, those who achieve that goal have been long-term practitioners of the art and are very deserving.
Horses were once a more important part of agriculture and highly selected to provide power, draft and transportation. With the advent of the modern combustion engine and other energy sources, the horse has become more important for recreation and unimportant as a source of power for industry and agriculture.
Although the horse does not contribute to the ‘food and fiber’ mission of agriculture, the horse industry still has a large impact on the economy. The results of a recent study by the American Horse Council (AHC, 2005) can be seen in Table 1.
Table 1. Economic impact of the horse industry in the United States.

The horse industry provides jobs and money to the American economy and has a comparable impact worldwide. Management practices and research that make the horse business more effective benefit our economy. Horse owners value horses that are athletic, healthy, intelligent and responsive.
Genetic selection and effective management are valuable tools; and genomics research on horses can enhance both selection and management. Genomics is transforming the way that we do research in agriculture. (Here, genomics is being used in a broad sense to include genomics, transcriptomics, proteomics, metabalomics, etc.)
Management practices and the foodstuff we provide our livestock influence the array of genes that are expressed throughout the tissues of the horse, even without consideration of hereditary potential.
Why agriculturalists support animal genomics
During the last 100 years, geneticists have accomplished amazing feats in food animal agriculture through genetic selection. Within the last 50 years alone, dairy cattle have doubled milk yields because of selection. Using the record-keeping model used by dairymen, meat producers made significant advances with beef, pork and poultry during the same period. Of course, these accomplishments were made without regard to DNA.
However, agricultural scientists are pragmatic if nothing else. In the late 1980s, a quantitative geneticist named Morris Soller projected the gains that one might realistically anticipate for the next 25 years using traditional quantitative genetics breeding methods for milk production.
Next, he projected the gain that one might obtain under the best circumstances following molecular biologists’ discovery of a gene with a major effect on production and consequent selection over the same 25 year period.
The results: quantitative genetics would win, no question. Still, this eminent quantitative geneticist advocated pursuing molecular genetics studies of production traits in livestock. Why? Because he also projected that one day the gains made using quantitative genetics would peak and that future advances would require an understanding of the molecular mechanisms underlying hereditary traits.
A strategy and rationale for applying markerassisted selection was described in a subsequent publication (Weller et al., 1990). Future advances in production would come through a comprehensive understanding of molecular genetics, a field now referred to as genomics.
Thumbnail history of the human genome sequencing projects
Genomics came to the fore of popular knowledge through the Human Genome Project (HGP). In October 1990 an international plan was established to fully sequence the human genome over a 15 year period. It was a bold plan, because at the time the technology did not exist to sequence and assemble a large complex genome.
Indeed, much of the funding and work was directed at developing the technologies. In May 1998, a private company (Celera) was formed to compete with the international effort and complete the human genome ahead of the HGP schedule. Theirs was a bold plan because it relied on computing power and computer programs that did not exist at the time.
Ultimately the two programs collaborated and announced a tie finish in the spring of 2003, coincidental with the 50th anniversary of Watson and Crick’s report on the structure of DNA. The applications of the information appear almost daily in our newspapers and have revolutionized medicine. The newly invented technologies also had a large impact in other arenas including agriculture and environmental sciences. We entered the ‘biology century’ (http://www.ornl.gov/sci/techresources/Human_Genome/ home.shtml).
History of animal genome projects
The applications in animal agriculture began in the early 1990s. In 1992, the United States Department of Agriculture (USDA) established the National Research Sponsored Project (NRSP) No. 8 in which monies were taken directly out of state appropriations for support of agricultural colleges to develop genomic studies for agriculturally important animals.
Scientists from land grant institutions in the United States along with scientists from industry and from around the world collaborated to create gene maps for livestock. The idea was to identify genes having a major effect on production traits or having a major impact on health and welfare of the animals.
From 1992 to 1997, NRSP8 included cattle, sheep, pigs and chickens. By 1997, scientists organized workshops on horse genomics and joined NRSP8 while aquaculture scientists joined NRSP8 in 2003. NRSP8 provides funds to coordinate collaborative research and meetings are held annually to discuss research advances and make plans for future collaborations.
NRSP8 has been one of the most successful collaborative efforts in agricultural research; effective gene maps have been created for all species and molecular determinants of health and production identified and diagnostic tests developed.
SEQUENCING THE GENOMES OF LIVESTOCK SPECIES
With completion of the human genome project, resources became available to sequence the genomes of other animal species. So far, among agriculturally important species we have completed genome sequences for chickens and cattle. Sequencing is underway for sheep and pigs. The initial projection for the human genome project was $3 billion.
However, that estimate included invention of technologies. The Celera project to sequence the human genome in 1998 was projected at a mere $300 million. Likewise, they had to invent technologies. The cost for sequencing the chicken genome was $60 million.
Cattle genome sequencing a few years later cost $52 million. Today, a low level sequencing is being done for pigs and sheep that identify only 85-90% of the genes at a cost of $10 million. Costs are going down and the assembly of the genomes is becoming easier as more species are sequenced and genome order assembled.
Genome organization is very similar among vertebrate species. Indeed, one of the major programs for sequencing the genomes of mammalian species is that of the National Human Genome Research Institute (NHGRI) (http://www.genome.gov/12511814). NHGRI has a program to sequence the genomes of diverse species in connection with annotation of the human genome.
In that program they are conducting low level DNA sequencing for diverse species including mice, rats, dogs, elephants, tree shrews, marsupials, tenrecs, cats, armadillos, rabbits, hedgehogs and guinea pigs.
As of February 2006, the horse has been added to that list. Comparison of the genome sequences of these different species helps to identify conserved regulatory regions in the DNA. These regulatory regions could not be found otherwise and the relevance of the large stretches of repetitive DNA sequences remains mysterious.
What we know about mammalian genomes
The genomics revolution revealed that all mammals possess approximately 20,000 pairs of genes. Within a cell, 10,000 of those genes will be expressed. The particular combination of 10,000 genes is characteristic of the tissue and its metabolic state. Genes may be up-regulated or down-regulated.
Investigation of gene expression is an area of genomics called functional genomics or transcriptomics. Gene expression is commonly assayed by Northern blots or by use of microarray assays allowing simultaneous evaluation of all possible genes. At the same time, genes have an intron-exon structure that allows expression of several structural versions.
Curiously, these 20,000 genes represent only 3% of the DNA in a cell. The remainder (97%) has been called ‘junk DNA’, but more because of our ignorance than a qualitative evaluation of the DNA.
Indeed, comparisons of DNA among diverse species reveals that vast stretches of DNA have been conserved over 100 million years of evolution (poultry to humans) and probably represent important regulatory DNA elements. This DNA is less conserved than genes but more conserved than random expectations.
Another surprising discovery was the conservation of gene order among vertebrates. Consequently, we can use the gene order in one species to predict the order in another. When we map a trait to a particular chromosome region and know its homology to the human genome organization, we can use the human genome sequence to predict candidate genes in livestock. Identifying candidate genes is really the first step in identifying gene mutations.
The low number of genes and their conservation among species is useful knowledge. The consequence of this observation for agriculture is that when a gene is found to cause a disease in humans, a similar disease in livestock is likely to be the product of a homologous gene.
We can use the information from human genomics to predict which genes are going to have an effect in livestock. For horses, this approach led to the discovery of the cause of hyperkalemic periodic paralysis (HYPP) in Quarter horses (Rudolph et al., 1992), severe combined immunodeficiency disease (SCID) in Arabian horses (Shin et al., 1997) and overo lethal white foal syndrome (OLWFS) in Paint horses (Metallinos et al., 1998; Santschi et al., 1998; Yang et al., 1998).
Genomics and horses
In 1995 a workshop was formed among scientists from among 25 laboratories working on problems related to genetics in horses (http://www.uky.edu/AG/horsemap). No one laboratory had all the resources necessary to complete a gene map for the horse; and this was the tool that each of the scientists needed to pursue their research into musculoskeletal diseases, infectious diseases, hereditary diseases, behavior, performance and even coat color patterns.
Indeed, by the 1990s it was clear that biological research on an organism would require a comprehensive gene map (By 2005 that requirement became a whole genome sequence!).
Over the next decade an agreement was reached on nomenclature for chromosomes (ISCNH, 1997), several linkage maps were reported (Lindgren et al., 1998; Guerin et al. 1999; Swinburne et al., 2000; Penedo et al., 2005; Swinburne et al., 2006), a radiation hybrid map was published (Chowdhary et al., 2003), and numerous other independent research projects filled in gaps, corrected technical errors and increased the efficacy of the horse gene map for identifying hereditary traits.
By 2006, 1000- 1500 genetic markers have been linkage mapped; more than 1200 genes have been mapped using cytogenetics and radiation hybrid techniques and we anticipate publication of a new radiation hybrid map within the next year that includes more than 4000 genes and genetic markers (Chowdhary, personal communication, January 2006).
As the map developed, it was used to identify genes and chromosome regions encoding hereditary traits in horses. The initial focus of the research efforts has been on simple Mendelian traits. The list of traits that have been mapped is shown in Table 2.
Table 2.Hereditary traits identified or mapped for the horse.


Discovering the genes for the traits listed in Table 2 represents the horse genetics equivalent of ‘harvesting the low-hanging fruit’. Each of the traits was clearly hereditary in nature.
Besides creating diagnostic tests for the traits, the most important aspects of these studies were to demonstrate that using horse genomics information could be effective to better characterize hereditary traits.
However, we need more powerful genomics tools to understand and resolve management questions for more complex traits in horses. For example, we anticipate that some musculoskeletal diseases of horses will involve multiple genetic systems, nutritional factors and management. Understanding such traits will require expertise from several disciplines. Indeed, most of the traits valued by horse breeders and horse owners are complex traits that combine selection, nutrition, training and other aspects of management.
Horses are valued for athleticism of event horses, strength of draft horses, strength and size of draft ponies, temperament, gait and, of course, racing performance of Thoroughbred horses.
The horse genome will be sequenced and assembled during 2006 by the NHGRI. The additional power from this information will have a dramatic impact on genomics tools. We can anticipate the following developments in the arena of genomics.
INVESTIGATIONS OF GENE EXPRESSION (TRANSCRIPTOMICS)
Currently, we have equine cDNA microarrays in development that allow us to assess 8,000 of the 20,000 horse genes.
The genome sequence will make it possible to create oligonucleotide arrays that assay expression of all 20,000 genes based on having the entire genome sequence. This means that nutritional studies can address the effect of nutrients on biochemical pathways or single genes.
As a horse is trained, as a horse recovers from disease and as a horse is inherently constituted, a unique set of genes will be expressed in its cells. Assaying the particular set of genes being expressed may provide valuable information for making choices about management, training and even selection.
The 20,000 element microarray will be a complex research tool, but as we use it to better understand nutrition and disease processes, this research tool will give way to many diagnostic tools.
GENETIC MARKER STUDIES
Currently we have a gene map with 1000 to 1500 genetic markers. Genetic studies rely on the use of several hundred mapped microsatellite markers that are segregated into families. The whole genome sequence will make it possible for us to assay tens of thousands of genetic markers called single nucleotide polymorphisms (SNPs).
Based on the large number of markers and with knowledge of the breed history, we can assay affected and unaffected individuals for a pattern of genetic variation at the population level.
This will be a particular boon because horse breeders may be reluctant to continue breeding horses suspected of having hereditary predispositions to disease. Families segregating for some traits are hard to find so we need to develop a strategy independent of family structure.
| Conclusions These tools will allow us to assess the way to use combinations of genetic selection, nutrition, management and training to produce the horses we need for our diverse purposes. In summary, I do wear a genetics hat. I am absolutely thrilled with the raw knowledge that has been produced about genomes during the last 20 years. Those of us in academia too often subscribe to the following sentiment: “It is indeed fortunate that God designed the universe along the precise lines of the academic disciplines.” (J. G. Miller). Nevertheless, our future successes and effective service to agriculture will depend on our ability to work across disciplinary lines. |
References
AHC. 2005. Horsepower. American Horse Council, Washington, DC.
Bailey, E., R.C. Reid, L.C. Skow, K. Mathiason, T.L. Lear and T.C. McGuire. 1997. Linkage of the gene for equine combined immunodeficiency disease to microsatellite markers HTG8 and HTG4; Synteny and FISH mapping to ECA9. Anim. Genetics 28:268-273.
Brooks, S.A., R.B. Terry and E. Bailey. 2002. Association of PCR-RFLP for KIT with Tobiano coat colour in horses. Anim. Genetics 33:301-303.
Brooks, S.A. and E. Bailey. 2005. Exon skipping in the KIT gene causes the Sabino spotting pattern in horses. Mammalian Genome 16:893-902.
Chowdhary, B.P., T. Raudsepp, S.R. Kata, G. Goh, L.V. Millon, V. Allan, F. Piumi, G. Guerin, J. Swinburne, M. Binns, T.L. Lear, J. Mickelson, J. Murray, D.F. Antczak, J.E. Womack and L.C. Skow. 2003. The first generation whole genome radiation hybrid map in the horse identifies conserved segments in human and mouse genomes. Genome Res. 13:742-751.
Guérin, G., E. Bailey, D. Bernoco, I. Anderson, D.F. Antczak, K. Bell, M.M. Binns, A.T. Bowling, R. Brandon, G. Cholewinski, E.G. Cothran, H. Ellegren, M. Förster, S. Godard, P. Horin, M. Ketchum, G. Lindgren, H. McPartlan, J-C. Mériaux, J.R. J.R Mickelson, L.V. Millon, J. Murray, A. Neau, K. Røed, K. Sandberg, Y-L. Shiue, L.C. Skow, M. Stott, J. Swinburne, S.J. Valberg, H. Van Haeringen, W.A. Van Haeringen and J. Zeigle. 1999. Report of the International Equine Gene Mapping Workshop: Male Linkage Map. Anim. Genetics 30:341-354.
Henner, J., P.A. Poncet, G. Guerin, C. Hagger, G. Stranzinger and S. Rieder. 2002. Genetic mapping of the(G)-locus responsible for the coat color phenotype ‘progressive graying with age’ in horses (Equus caballus). Mammal Genome 13:535-537.
ISCNH. 1997. International system for cytogenetic nomenclature of the domestic horse. A.T. Bowling, M. Breen, B.P. Chowdhary, K. Hirota, T.L. Lear, L.V. Millon, F.A. Ponce de Leon, T. Raudsepp and G. Stranzinger (Committee). Chrom. Res. 5:443- 453.
Lindgren, G., K. Sandberg, H. Persson, S. Marklund, M. Breen, B. Sandgren, J. Carlstén and H. Ellegren. 1998. A primary male autosomal linkage map of the horse genome. Genome Res. 8:951-966.
Locke, M.M., M.C.T. Penedo, S.J. Bricker, L.V. Millon and J.D. Murray. 2002. Linkage of the gray coat color locus to microsatellites on horse chromosome 25. Anim. Genetics 33:329-337.
Locke, M.M., L.S. Ruth, L.V. Millon, M.C.T. Penedo, J.D. Murray and A.T. Bowling. 2001. The cream dilution gene, responsible for the palomino and buckskin coat colors, maps to horse chromosome 21. Anim. Genetics 32:340-343.
Mariat, D., S. Taourit and G. Guerin. 2003 A mutation in the MATP gene causes the cream coat color in the horse. Genet. Sel. Evol. 35:119-133.
Marklund, L., M. Johanssonn Moller, K. Sandberg and L. Andersson. 1996. A missense mutation in the gene for melanocyte-stimulating homone receptor (MC1R) is associated with the chestnut coat color in horses. Mammal. Genome 7:895-899.
Mau, C., P.A. Poncet, B. Bucher, G. Stranzinger and W. Rieder. 2004. Genetic mapping of dominant white (W) a homozygous lethal condition in the horse (Equus caballus). Animal Breeding and Genetics 121:374-383.
Metallinos, D.L., A.T. Bowling and J. Rine. 1998. A missense mutation in the endothelin- β receptor gene is associated with lethal white foal syndrome: an equine version of Hirschsprung Disease. Mammal. Genome 9:426-431.
Milenkovic, D., A. Chaffaux, S. Taourit and G. Guerin. 2003. A mutation in the LAMC2 gene causes the Herlitz junctional epidermolysis bullosa (H-JEB) in two French draft horse breeds. Genetic Selection and Evolution 35:249-256.
Penedo, M.C.T., L.V. Millon, D. Bernoco, E. Bailey, M.M. Binns, G. Cholewinski, N. Ellis, J. Flynn, B. Gralak, A. Guthrie, G. Lindgren, L.A. Lyons, T. Tozaki, K. Røed and J. Swinburne. 2005. International Equine Gene Mapping Workshop Report: A comprehensive linkage map constructed with data from new markers and by merging four mapping resources. Cytogenet. Genome Res. 111:5-15.
Rieder, S., S. Taourit, D. Mariat, B. Langlois and G. Guerin. 2001. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mammal. Genome 12:450-455.
Rudolph, J., S. Spier, G. Byrns, C. Rojas, D. Bernoco and E. Hoffman. 1992. Periodic paralysis in Quarter Horses, a sodium channel mutation disseminated by selective breeding. Nat. Genetics 2:144-147.
Santschi, E.M., A.K. Purdy, S.J. Valberg, P.D. Vrotsos, H. Kaese and J.R. Mickelson. 1998. Endothelin receptor β polymorphism associated with lethal white foal syndrome in horses. Mammal. Genome 9:306-309.
Shin, E.K., L.E. Perryman and K. Meek. 1997. A kinase negative mutation of DNAPKcs in equine SCID results in defective coding and signal joint formation. J. Immunol. 158:3565-3569.
Spirito, F., A. Charlesworth, K. Linder, J.P. Ortonne, J. Baird and G. Meneguzzi. 2002. Animal models for skin blistering conditions: absence of laminin 5 causes hereditary junctional mechanobullous disease in the Belgian horse. J. Invest. Dermatol. 119:684- 91.
Swinburne, J.E. M. Boursnell, G. Hill, L. Pettitt, T. Allen, B. Chowdhary, T. Hasegawa, M. Kurosawa, T. Leeb, S. Mashima, J.R. Mickelson, T. Raudsepp, T. Tozaki and M. Binns. 2006. Single linkage group per chromosome genetic linkage map for the horse, based on two three-generation, full-sibling, crossbred horse reference families. Genomics 87:1-29.
Swinburne, J., C. Gerstenberg, M. Breen, V. Aldridge, L. Lockhart, E. Marti, D.F. Antczak, M. Egglestone-Stott, E. Bailey, J. Mickelson, K. Röed, G. Lindgren, W. van Haeringen, G. Guérin, J. Bjornasson, W.R. Allen and M.M. Binns. 2000. First comprehensive low-density horse linkage map based on two, three-generation, fullsibling, cross-bred horse reference families. Genomics 66:123-134.
Swinburne, J.E., A. Hopkins and M.M. Binns. 2002. Assignment of the horse grey color gene to ECA25 using whole genome scanning. Anim. Genetics 33:338-342.
Terry, R.B., S. Archer, S. Brooks, D. Bernoco and E. Bailey. 2004. Linkage mapping the appaloosa coat color gene (LP) of the horse. Animal Genetics 35:134-137.
Ward, T.L., S.H.J. Valberg, D.L. Adelson, C.A. Abbey, M.M. Binns and J.R. Mickelson. 2004. Glycogen branching enzyme (GBE1) mutation causing equine glycogen storage disease IV. Mammal. Genome 15:570-577.
Weller, J.L., Y. Kashi and M. Soller. 1990. Power of daughter and granddaughter designs for determining linkage between marker loci and quantitative trait loci in dairy cattle. J. Dairy Sci. 73:2525-2537.
Yang, G.C., D. Croaker, A.L. Zhang, P. Manglick, T. Cartmill and D. Cass. 1998. A dinucleotide mutation in the endothelin-B receptor gene is associated with lethal while foal syndrome (LWFS) a horse variant of Hirschspring disease. Human Molecular Genetics 7:1047-1052.
Author: ERNEST BAILEY
M.H. Gluck Equine Research Center, University of Kentucky, Lexington, Kentucky, USA
M.H. Gluck Equine Research Center, University of Kentucky, Lexington, Kentucky, USA
Related topics
Join to be able to comment.
Once you join Engormix, you will be able to participate in all content and forums.
* Required information
Would you like to discuss another topic? Create a new post to engage with experts in the community.
Create a post